Assembly of lipopolysaccharide in Escherichia coli requires the essential LapB heat shock protein
- PMID: 24722986
- PMCID: PMC4031536
- DOI: 10.1074/jbc.M113.539494
Assembly of lipopolysaccharide in Escherichia coli requires the essential LapB heat shock protein
Abstract
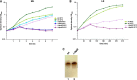
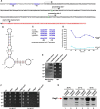
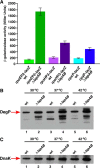
Here, we describe two new heat shock proteins involved in the assembly of LPS in Escherichia coli, LapA and LapB (lipopolysaccharide assembly protein A and B). lapB mutants were identified based on an increased envelope stress response. Envelope stress-responsive pathways control key steps in LPS biogenesis and respond to defects in the LPS assembly. Accordingly, the LPS content in ΔlapB or Δ(lapA lapB) mutants was elevated, with an enrichment of LPS derivatives with truncations in the core region, some of which were pentaacylated and exhibited carbon chain polymorphism. Further, the levels of LpxC, the enzyme that catalyzes the first committed step of lipid A synthesis, were highly elevated in the Δ(lapA lapB) mutant. Δ(lapA lapB) mutant accumulated extragenic suppressors that mapped either to lpxC, waaC, and gmhA, or to the waaQ operon (LPS biosynthesis) and lpp (Braun's lipoprotein). Increased synthesis of either FabZ (3-R-hydroxymyristoyl acyl carrier protein dehydratase), slrA (novel RpoE-regulated non-coding sRNA), lipoprotein YceK, toxin HicA, or MurA (UDP-N-acetylglucosamine 1-carboxyvinyltransferase) suppressed some of the Δ(lapA lapB) defects. LapB contains six tetratricopeptide repeats and, at the C-terminal end, a rubredoxin-like domain that was found to be essential for its activity. In pull-down experiments, LapA and LapB co-purified with LPS, Lpt proteins, FtsH (protease), DnaK, and DnaJ (chaperones). A specific interaction was also observed between WaaC and LapB. Our data suggest that LapB coordinates assembly of proteins involved in LPS synthesis at the plasma membrane and regulates turnover of LpxC, thereby ensuring balanced biosynthesis of LPS and phospholipids consistent with its essentiality.
Keywords: Endotoxin; Glycobiology; Glycolipid Structure; Glycosyltransferases; Heptosyltransferase; Lipid A; Lipopolysaccharide (LPS); Myristoyltransferase; RpoH; Tetratricopeptide Repeat (TPR).
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures









References
-
- Gronow S., Brade H. (2001) Lipopolysaccharide biosynthesis: which steps do bacteria need to survive? J. Endotoxin Res. 7, 3–23 - PubMed
-
- Frirdich E., Whitfield C. (2005) Lipopolysaccharide inner core oligosaccharide structure and outer membrane stability in human pathogens belonging to the Enterobacteriaceae. J. Endotoxin Res. 11, 133–144 - PubMed
-
- Klein G., Lindner B., Brade H., Raina S. (2011) Molecular basis of lipopolysaccharide heterogeneity in Escherichia coli: envelope stress-responsive regulators control the incorporation of glycoforms with a third 3-deoxy-α-d-manno-oct-2-ulosonic acid and rhamnose. J. Biol. Chem. 286, 42787–42807 - PMC - PubMed
-
- Clementz T., Raetz C. R. (1991) A gene coding for 3-deoxy-d-manno-octulosonic-acid transferase in Escherichia coli: identification, mapping, cloning, and sequencing. J. Biol. Chem. 266, 9687–9696 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

