What's that gene (or protein)? Online resources for exploring functions of genes, transcripts, and proteins
- PMID: 24723265
- PMCID: PMC3982986
- DOI: 10.1091/mbc.E13-10-0602
What's that gene (or protein)? Online resources for exploring functions of genes, transcripts, and proteins
Abstract
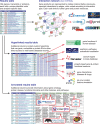
The genomic era has enabled research projects that use approaches including genome-scale screens, microarray analysis, next-generation sequencing, and mass spectrometry-based proteomics to discover genes and proteins involved in biological processes. Such methods generate data sets of gene, transcript, or protein hits that researchers wish to explore to understand their properties and functions and thus their possible roles in biological systems of interest. Recent years have seen a profusion of Internet-based resources to aid this process. This review takes the viewpoint of the curious biologist wishing to explore the properties of protein-coding genes and their products, identified using genome-based technologies. Ten key questions are asked about each hit, addressing functions, phenotypes, expression, evolutionary conservation, disease association, protein structure, interactors, posttranslational modifications, and inhibitors. Answers are provided by presenting the latest publicly available resources, together with methods for hit-specific and data set-wide information retrieval, suited to any genome-based analytical technique and experimental species. The utility of these resources is demonstrated for 20 factors regulating cell proliferation. Results obtained using some of these are discussed in more depth using the p53 tumor suppressor as an example. This flexible and universally applicable approach for characterizing experimental hits helps researchers to maximize the potential of their projects for biological discovery.
Figures

Similar articles
-
Managing Sequence Data.Methods Mol Biol. 2017;1525:79-106. doi: 10.1007/978-1-4939-6622-6_4. Methods Mol Biol. 2017. PMID: 27896718
-
Genomic Database Searching.Methods Mol Biol. 2017;1525:225-269. doi: 10.1007/978-1-4939-6622-6_10. Methods Mol Biol. 2017. PMID: 27896724
-
A web application for the unspecific detection of differentially expressed DNA regions in strand-specific expression data.Bioinformatics. 2015 Oct 1;31(19):3228-30. doi: 10.1093/bioinformatics/btv343. Epub 2015 Jun 2. Bioinformatics. 2015. PMID: 26040457
-
Locus-specific databases in cancer: what future in a post-genomic era? The TP53 LSDB paradigm.Hum Mutat. 2014 Jun;35(6):643-53. doi: 10.1002/humu.22518. Epub 2014 Mar 5. Hum Mutat. 2014. PMID: 24478183 Review.
-
The humankind genome: from genetic diversity to the origin of human diseases.Genome. 2013 Dec;56(12):705-16. doi: 10.1139/gen-2013-0125. Genome. 2013. PMID: 24433206 Review.
Cited by
-
Only one health, and so many omics.Cancer Cell Int. 2015 Jun 23;15:64. doi: 10.1186/s12935-015-0212-2. eCollection 2015. Cancer Cell Int. 2015. PMID: 26101467 Free PMC article.
-
Protein Sequence Annotation Tool (PSAT): a centralized web-based meta-server for high-throughput sequence annotations.BMC Bioinformatics. 2016 Jan 20;17:43. doi: 10.1186/s12859-016-0887-y. BMC Bioinformatics. 2016. PMID: 26792120 Free PMC article.
-
Differential gene expression profile of multinodular goiter.PLoS One. 2022 May 20;17(5):e0268354. doi: 10.1371/journal.pone.0268354. eCollection 2022. PLoS One. 2022. PMID: 35594253 Free PMC article.
-
Effect of Tension on Human Periodontal Ligament Cells: Systematic Review and Network Analysis.Front Bioeng Biotechnol. 2021 Aug 27;9:695053. doi: 10.3389/fbioe.2021.695053. eCollection 2021. Front Bioeng Biotechnol. 2021. PMID: 34513810 Free PMC article.
-
Genomic association with pathogen carriage in bighorn sheep (Ovis canadensis).Ecol Evol. 2021 Mar 2;11(6):2488-2502. doi: 10.1002/ece3.7159. eCollection 2021 Mar. Ecol Evol. 2021. PMID: 33767816 Free PMC article.
References
-
- Alberts B. The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell. 1998;92:291–294. - PubMed
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Amberger J, Bocchini C, Hamosh A. A new face and new challenges for Online Mendelian Inheritance in Man (OMIM) Hum Mutat. 2011;32:564–567. - PubMed
-
- Asplund A, Edqvist PH, Schwenk JM, Ponten F. Antibodies for profiling the human proteome—The Human Protein Atlas as a resource for cancer research. Proteomics. 2012;12:2067–2077. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

