A web-portal for interactive data exploration, visualization, and hypothesis testing
- PMID: 24723882
- PMCID: PMC3972454
- DOI: 10.3389/fninf.2014.00025
A web-portal for interactive data exploration, visualization, and hypothesis testing
Abstract
Clinical research studies generate data that need to be shared and statistically analyzed by their participating institutions. The distributed nature of research and the different domains involved present major challenges to data sharing, exploration, and visualization. The Data Portal infrastructure was developed to support ongoing research in the areas of neurocognition, imaging, and genetics. Researchers benefit from the integration of data sources across domains, the explicit representation of knowledge from domain experts, and user interfaces providing convenient access to project specific data resources and algorithms. The system provides an interactive approach to statistical analysis, data mining, and hypothesis testing over the lifetime of a study and fulfills a mandate of public sharing by integrating data sharing into a system built for active data exploration. The web-based platform removes barriers for research and supports the ongoing exploration of data.
Keywords: data dictionary; data exploration; data sharing; genetics; hypothesis testing; imaging.
Figures

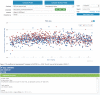

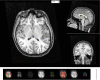




References
-
- Akaike H. (1974). A new look at the statistical model identification. IEEE Trans. Automat. Contr. 19, 716–723 10.1109/TAC.1974.1100705 - DOI
-
- ASM. (2013). ASM. Availble online at: http://asmjs.org/, Last viewed July 2013.
-
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Statist. Soc. Ser. B 57, 289–300
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

