Intracellular coordination of potyviral RNA functions in infection
- PMID: 24723931
- PMCID: PMC3972461
- DOI: 10.3389/fpls.2014.00110
Intracellular coordination of potyviral RNA functions in infection
Abstract
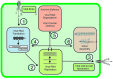
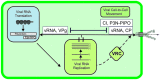
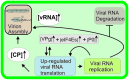
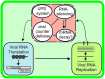
Establishment of an infection cycle requires mechanisms to allocate the genomes of (+)-stranded RNA viruses in a balanced ratio to translation, replication, encapsidation, and movement, as well as mechanisms to prevent translocation of viral RNA (vRNA) to cellular RNA degradation pathways. The ratio of vRNA allocated to various functions is likely balanced by the availability of regulatory proteins or competition of the interaction sites within regulatory ribonucleoprotein complexes. Due to the transient nature of viral processes and the interdependency between vRNA pathways, it is technically demanding to work out the exact molecular mechanisms underlying vRNA regulation. A substantial number of viral and host proteins have been identified that facilitate the steps that lead to the assembly of a functional potyviral RNA replication complex and their fusion with chloroplasts. Simultaneously with on-going viral replication, part of the replicated potyviral RNA enters movement pathways. Although not much is known about the processes of potyviral RNA release from viral replication complexes, the molecular interactions involved in these processes determine the fate of the replicated vRNA. Some viral and host cell proteins have been described that direct replicated potyviral RNA to translation to enable potyviral gene expression and productive infection. The antiviral defense of the cell causes vRNA degradation by RNA silencing. We hypothesize that also plant pathways involved in mRNA decay may have a role in the coordination of potyviral RNA expression. In this review, we discuss the roles of different potyviral and host proteins in the coordination of various potyviral RNA functions.
Keywords: potyviral RNA degradation; potyviral RNA encapsidation; potyviral RNA functions; potyviral movement; potyviral replication; potyviral translation; potyviruses.
Figures
References
-
- Abdul-Razzak A., Guiraud T., Peypelut M., Walter J., Houvenaghel M. C., Candresse T., et al. (2009). Involvement of the cylindrical inclusion (CI) protein in the overcoming of an eIF4E-mediated resistance against lettuce mosaic Potyvirus. Mol. Plant Pathol. 10 109–113 10.1111/j.1364-3703.2008.00513.x - DOI - PMC - PubMed
-
- Ala-Poikela M., Goytia E., Haikonen T., Rajamäki M.-L, Valkonen J. P. T. (2011). Helper component proteinase of the genus Potyvirus is an interaction partner of translation initiation factors eIF(iso)4E and eIF4E and contains a 4E binding motif. J. Virol. 85 6784–6794 10.1128/JVI.00485-11 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

