Commonalities and differences of T3SSs in rhizobia and plant pathogenic bacteria
- PMID: 24723933
- PMCID: PMC3973906
- DOI: 10.3389/fpls.2014.00114
Commonalities and differences of T3SSs in rhizobia and plant pathogenic bacteria
Abstract
Plant pathogenic bacteria and rhizobia infect higher plants albeit the interactions with their hosts are principally distinct and lead to completely different phenotypic outcomes, either pathogenic or mutualistic, respectively. Bacterial protein delivery to plant host plays an essential role in determining the phenotypic outcome of plant-bacteria interactions. The involvement of type III secretion systems (T3SSs) in mediating animal- and plant-pathogen interactions was discovered in the mid-80's and is now recognized as a multiprotein nanomachine dedicated to trans-kingdom movement of effector proteins. The discovery of T3SS in bacteria with symbiotic lifestyles broadened its role beyond virulence. In most T3SS-positive bacterial pathogens, virulence is largely dependent on functional T3SSs, while in rhizobia the system is dispensable for nodulation and can affect positively or negatively the mutualistic associations with their hosts. This review focuses on recent comparative genome analyses in plant pathogens and rhizobia that uncovered similarities and variations among T3SSs in their genetic organization, regulatory networks and type III secreted proteins and discusses the evolutionary adaptations of T3SSs and type III secreted proteins that might account for the distinguishable phenotypes and host range characteristics of plant pathogens and symbionts.
Keywords: atypical T3SSs; nodulation; pili; plant pathogenic bacteria; plant-associated bacteria; rhizobia; translocator; type III secretion.
Figures
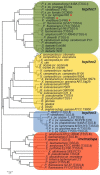
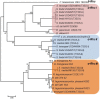

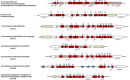
References
-
- Alavi S. M., Sanjari S., Durand F., Brin C., Manceau C., Poussier S. (2008). Assessment of the genetic diversity of Xanthomonas axonopodis pv. phaseoli and Xanthomonas fuscans subsp. fuscans as a basis to identify putative pathogenicity genes and a type III secretion system of the SPI-1 family by multiple suppression subtractive hybridizations. Appl. Environ. Microbiol. 74, 3295–3301 10.1128/AEM.02507-07 - DOI - PMC - PubMed
-
- Alfano J. R., Charkowski A. O., Deng W. L., Badel J. L., Petnicki-Ocwieja T., van Dijk K., et al. (2000). The Pseudomonas syringae Hrp pathogenicity island has a tripartite mosaic structure composed of a cluster of type III secretion genes bounded by exchangeable effector and conserved effector loci that contribute to parasitic fitness and pathogenicity in plants. Proc. Natl. Acad. Sci. U.S.A. 97, 4856–4861 10.1073/pnas.97.9.4856 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

