Asymmetric friction of nonmotor MAPs can lead to their directional motion in active microtubule networks
- PMID: 24725408
- PMCID: PMC4015189
- DOI: 10.1016/j.cell.2014.02.018
Asymmetric friction of nonmotor MAPs can lead to their directional motion in active microtubule networks
Abstract
Diverse cellular processes require microtubules to be organized into distinct structures, such as asters or bundles. Within these dynamic motifs, microtubule-associated proteins (MAPs) are frequently under load, but how force modulates these proteins' function is poorly understood. Here, we combine optical trapping with TIRF-based microscopy to measure the force dependence of microtubule interaction for three nonmotor MAPs (NuMA, PRC1, and EB1) required for cell division. We find that frictional forces increase nonlinearly with MAP velocity across microtubules and depend on filament polarity, with NuMA's friction being lower when moving toward minus ends, EB1's lower toward plus ends, and PRC1's exhibiting no directional preference. Mathematical models predict, and experiments confirm, that MAPs with asymmetric friction can move directionally within actively moving microtubule pairs they crosslink. Our findings reveal how nonmotor MAPs can generate frictional resistance in dynamic cytoskeletal networks via micromechanical adaptations whose anisotropy may be optimized for MAP localization and function within cellular structures.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures
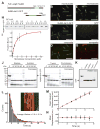

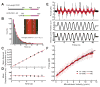
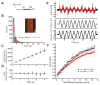

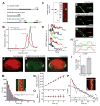

Comment in
-
Friction on MAP determines its traveling direction on microtubules.Dev Cell. 2014 Apr 14;29(1):5-6. doi: 10.1016/j.devcel.2014.03.022. Dev Cell. 2014. PMID: 24735876
References
-
- Bieling P, Laan L, Schek H, Munteanu EL, Sandblad L, Dogterom M, Brunner D, Surrey T. Reconstitution of a microtubule plus-end tracking system in vitro. Nature. 2007;450:1100–1105. - PubMed
-
- Bieling P, Telley IA, Surrey T. A Minimal Midzone Protein Module Controls Formation and Length of Antiparallel Microtubule Overlaps. Cell. 2010;142:420–432. - PubMed
-
- Bormuth V, Varga V, Howard J, Schaffer E. Protein Friction Limits Diffusive and Directed Movements of Kinesin Motors on Microtubules. Science. 2009;325:870–873. - PubMed
-
- Braun M, Lansky Z, Fink G, Ruhnow F, Diez S, Janson ME. Adaptive braking by Ase1 prevents overlapping microtubules from sliding completely apart. Nat Cell Biol. 2011;13:1259–U1201. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

