dRiskKB: a large-scale disease-disease risk relationship knowledge base constructed from biomedical text
- PMID: 24725842
- PMCID: PMC3998061
- DOI: 10.1186/1471-2105-15-105
dRiskKB: a large-scale disease-disease risk relationship knowledge base constructed from biomedical text
Abstract
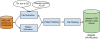
Background: Discerning the genetic contributions to complex human diseases is a challenging mandate that demands new types of data and calls for new avenues for advancing the state-of-the-art in computational approaches to uncovering disease etiology. Systems approaches to studying observable phenotypic relationships among diseases are emerging as an active area of research for both novel disease gene discovery and drug repositioning. Currently, systematic study of disease relationships on a phenome-wide scale is limited due to the lack of large-scale machine understandable disease phenotype relationship knowledge bases. Our study innovates a semi-supervised iterative pattern learning approach that is used to build an precise, large-scale disease-disease risk relationship (D1 → D2) knowledge base (dRiskKB) from a vast corpus of free-text published biomedical literature.
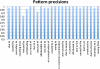
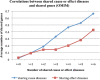
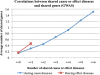
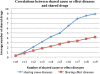
Results: 21,354,075 MEDLINE records comprised the text corpus under study. First, we used one typical disease risk-specific syntactic pattern (i.e. "D1 due to D2") as a seed to automatically discover other patterns specifying similar semantic relationships among diseases. We then extracted D1 → D2 risk pairs from MEDLINE using the learned patterns. We manually evaluated the precisions of the learned patterns and extracted pairs. Finally, we analyzed the correlations between disease-disease risk pairs and their associated genes and drugs. The newly created dRiskKB consists of a total of 34,448 unique D1 → D2 pairs, representing the risk-specific semantic relationships among 12,981 diseases with each disease linked to its associated genes and drugs. The identified patterns are highly precise (average precision of 0.99) in specifying the risk-specific relationships among diseases. The precisions of extracted pairs are 0.919 for those that are exactly matched and 0.988 for those that are partially matched. By comparing the iterative pattern approach starting from different seeds, we demonstrated that our algorithm is robust in terms of seed choice. We show that diseases and their risk diseases as well as diseases with similar risk profiles tend to share both genes and drugs.
Conclusions: This unique dRiskKB, when combined with existing phenotypic, genetic, and genomic datasets, can have profound implications in our deeper understanding of disease etiology and in drug repositioning.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

