The others: our biased perspective of eukaryotic genomes
- PMID: 24726347
- PMCID: PMC4342545
- DOI: 10.1016/j.tree.2014.03.006
The others: our biased perspective of eukaryotic genomes
Abstract
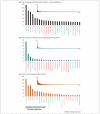
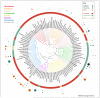
Understanding the origin and evolution of the eukaryotic cell and the full diversity of eukaryotes is relevant to many biological disciplines. However, our current understanding of eukaryotic genomes is extremely biased, leading to a skewed view of eukaryotic biology. We argue that a phylogeny-driven initiative to cover the full eukaryotic diversity is needed to overcome this bias. We encourage the community: (i) to sequence a representative of the neglected groups available at public culture collections, (ii) to increase our culturing efforts, and (iii) to embrace single cell genomics to access organisms refractory to propagation in culture. We hope that the community will welcome this proposal, explore the approaches suggested, and join efforts to sequence the full diversity of eukaryotes.
Keywords: culture collections; culturing bias; ecology; eukaryotic genomics; eukaryotic tree of life; phylogeny; single cell genomics.
Copyright © 2014 Elsevier Ltd. All rights reserved.
Figures


References
-
- Stanier RY, et al. The Microbial World. Prentice-Hall; 1957.
-
- Pennisi E. No genome left behind. Science. 2009;326:794–795. - PubMed
-
- Bennetzen J, Kellogg E. A plant genome initiative. Plant Cell. 1998;10:488–494.
-
- Galagan JE, et al. Genomics of the fungal kingdom: insights into eukaryotic biology. Genome Res. 2005;15:1620–1631. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

