Testing the role of myeloid cell glucose flux in inflammation and atherosclerosis
- PMID: 24726364
- PMCID: PMC4021396
- DOI: 10.1016/j.celrep.2014.03.028
Testing the role of myeloid cell glucose flux in inflammation and atherosclerosis
Abstract
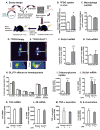
Inflammatory activation of myeloid cells is accompanied by increased glycolysis, which is required for the surge in cytokine production. Although in vitro studies suggest that increased macrophage glucose metabolism is sufficient for cytokine induction, the proinflammatory effects of increased myeloid cell glucose flux in vivo and the impact on atherosclerosis, a major complication of diabetes, are unknown. We therefore tested the hypothesis that increased glucose uptake in myeloid cells stimulates cytokine production and atherosclerosis. Overexpression of the glucose transporter GLUT1 in myeloid cells caused increased glycolysis and flux through the pentose phosphate pathway but did not induce cytokines. Moreover, myeloid-cell-specific overexpression of GLUT1 in LDL receptor-deficient mice was ineffective in promoting atherosclerosis. Thus, increased glucose flux is insufficient for inflammatory myeloid cell activation and atherogenesis. If glucose promotes atherosclerosis by increasing cellular glucose flux, myeloid cells do not appear to be the key targets.
Copyright © 2014 The Authors. Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Brown A, Reynolds LR, Bruemmer D. Intensive glycemic control and cardiovascular disease: an update. Nat Rev Cardiol. 2010;7:369–375. - PubMed
-
- Cline GW, Jucker BM, Trajanoski Z, Rennings AJ, Shulman GI. A novel 13C NMR method to assess intracellular glucose concentration in muscle, in vivo. Am J Physiol. 1998;274(2 Pt 1):E381–E389. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 HL018645/HL/NHLBI NIH HHS/United States
- R01 HL097365/HL/NHLBI NIH HHS/United States
- P01HL092969/HL/NHLBI NIH HHS/United States
- R01 HL067267/HL/NHLBI NIH HHS/United States
- R33DK069801/DK/NIDDK NIH HHS/United States
- P30DK035816/DK/NIDDK NIH HHS/United States
- R01HL081795/HL/NHLBI NIH HHS/United States
- P30 DK089503/DK/NIDDK NIH HHS/United States
- R01HL097365/HL/NHLBI NIH HHS/United States
- R01HL067267/HL/NHLBI NIH HHS/United States
- P30 DK020572/DK/NIDDK NIH HHS/United States
- U24 DK097153/DK/NIDDK NIH HHS/United States
- P30 DK017047/DK/NIDDK NIH HHS/United States
- R01 HL062887/HL/NHLBI NIH HHS/United States
- R33 DK069801/DK/NIDDK NIH HHS/United States
- P30DK017047/DK/NIDDK NIH HHS/United States
- P30 DK035816/DK/NIDDK NIH HHS/United States
- P30DK089503/DK/NIDDK NIH HHS/United States
- U24DK097153/DK/NIDDK NIH HHS/United States
- R01 HL081795/HL/NHLBI NIH HHS/United States
- P01HL018645/HL/NHLBI NIH HHS/United States
- R01HL062887/HL/NHLBI NIH HHS/United States
- P01 HL092969/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

