Targeting lactate dehydrogenase--a inhibits tumorigenesis and tumor progression in mouse models of lung cancer and impacts tumor-initiating cells
- PMID: 24726384
- PMCID: PMC4096909
- DOI: 10.1016/j.cmet.2014.03.003
Targeting lactate dehydrogenase--a inhibits tumorigenesis and tumor progression in mouse models of lung cancer and impacts tumor-initiating cells
Abstract
The lactate dehydrogenase-A (LDH-A) enzyme catalyzes the interconversion of pyruvate and lactate, is upregulated in human cancers, and is associated with aggressive tumor outcomes. Here we use an inducible murine model and demonstrate that inactivation of LDH-A in mouse models of NSCLC driven by oncogenic K-RAS or EGFR leads to decreased tumorigenesis and disease regression in established tumors. We also show that abrogation of LDH-A results in reprogramming of pyruvate metabolism, with decreased lactic fermentation in vitro, in vivo, and ex vivo. This was accompanied by reactivation of mitochondrial function in vitro, but not in vivo or ex vivo. Finally, using a specific small molecule LDH-A inhibitor, we demonstrated that LDH-A is essential for cancer-initiating cell survival and proliferation. Thus, LDH-A can be a viable therapeutic target for NSCLC, including cancer stem cell-dependent drug-resistant tumors.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures
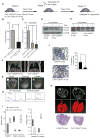
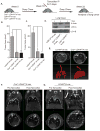
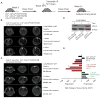
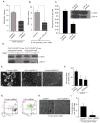

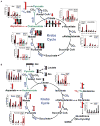
 ,
,
 : 13C in the first and second turn without PC input;
: 13C in the first and second turn without PC input;
 : 13C with PC input.
: 13C with PC input.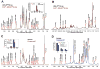
References
-
- Ailles LE, Weissman IL. Cancer stem cells in solid tumors. Curr Opin Biotechnol. 2007;18:460–466. - PubMed
-
- Amann J, Kalyankrishna S, Massion PP, Ohm JE, Girard L, Shigematsu H, Peyton M, Juroske D, Huang Y, Stuart Salmon J, et al. Aberrant epidermal growth factor receptor signaling and enhanced sensitivity to EGFR inhibitors in lung cancer. Cancer Res. 2005;65:226–235. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

