Fli1 acts downstream of Etv2 to govern cell survival and vascular homeostasis via positive autoregulation
- PMID: 24727028
- PMCID: PMC4080722
- DOI: 10.1161/CIRCRESAHA.1134303145
Fli1 acts downstream of Etv2 to govern cell survival and vascular homeostasis via positive autoregulation
Abstract
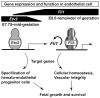
Rationale: Cardiovascular health depends on proper development and integrity of blood vessels. Ets variant 2 (Etv2), a member of the E26 transforming-specific family of transcription factors, is essential to initiate a transcriptional program leading to vascular morphogenesis in early mouse embryos. However, endothelial expression of the Etv2 gene ceases at midgestation; therefore, vascular development past this stage must continue independent of Etv2.
Objective: To identify molecular mechanisms underlying transcriptional regulation of vascular morphogenesis and homeostasis in the absence of Etv2.
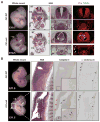
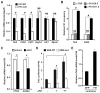
Methods and results: Using loss- and gain-of-function strategies and a series of molecular techniques, we identify Friend leukemia integration 1 (Fli1), another E26 transforming-specific family transcription factor, as a downstream target of Etv2. We demonstrate that Etv2 binds to conserved Ets-binding sites within the promoter region of the Fli1 gene and governs Fli1 expression. Importantly, in the absence of Etv2 at midgestation, binding of Etv2 at Ets-binding sites in the Fli1 promoter is replaced by Fli1 protein itself, sustaining expression of Fli1 as well as selective Etv2-regulated endothelial genes to promote endothelial cell survival and vascular integrity. Consistent with this, we report that Fli1 binds to the conserved Ets-binding sites within promoter and enhancer regions of other Etv2-regulated endothelial genes, including Tie2, to control their expression at and beyond midgestation.
Conclusions: We have identified a novel positive feed-forward regulatory loop in which Etv2 activates expression of genes involved in vasculogenesis, including Fli1. Once the program is activated in early embryos, Fli1 then takes over to sustain the process in the absence of Etv2.
Keywords: apoptosis; developmental biology; embryonic development; gene expression; homeostasis; transcription factors.
© 2014 American Heart Association, Inc.
Figures



References
-
- Olson EN, Srivastava D. Molecular pathways controlling heart development. Science. 1996;272:671–676. - PubMed
-
- Choi K. The hemangioblast: A common progenitor of hematopoietic and endothelial cells. J Hematother Stem Cell Res. 2002;11:91–101. - PubMed
-
- Chung YS, Zhang WJ, Arentson E, Kingsley PD, Palis J, Choi K. Lineage analysis of the hemangioblast as defined by Flk1 and Scl expression. Development. 2002;129:5511–5520. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

