Investigating the interactions of yeast prions: [SWI+], [PSI+], and [PIN+]
- PMID: 24727082
- PMCID: PMC4063924
- DOI: 10.1534/genetics.114.163402
Investigating the interactions of yeast prions: [SWI+], [PSI+], and [PIN+]
Abstract
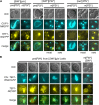
Multiple prion elements, which are transmitted as heritable protein conformations and often linked to distinct phenotypes, have been identified in the budding yeast, Saccharomyces cerevisiae. It has been shown that overproduction of a prion protein Swi1 can promote the de novo conversion of another yeast prion [PSI(+)] when Sup35 is co-overproduced. However, the mechanism underlying this Pin(+) ([PSI(+)] inducible) activity is not clear. Moreover, how the Swi1 prion ([SWI(+)]) interacts with other yeast prions is unknown. Here, we demonstrate that the Pin(+) activity associated with Swi1 overproduction is independent of Rnq1 expression or [PIN(+)] conversion. We also show that [SWI(+)] enhances the appearance of [PSI(+)] and [PIN(+)]. However, [SWI(+)] significantly compromises the Pin(+) activity of [PIN(+)] when they coexist. We further demonstrate that a single yeast cell can harbor three prions, [PSI(+)], [PIN(+)], and [SWI(+)], simultaneously. However, under this condition, [SWI(+)] is significantly destabilized. While the propensity to aggregate underlies prionogenesis, Swi1 and Rnq1 aggregates resulting from overproduction are usually nonheritable. Conversely, prion protein aggregates formed in nonoverexpressing conditions or induced by preexisting prion(s) are more prionogenic. For [PSI(+)] and [PIN(+)] de novo formation, heterologous "facilitators," such as preexisting [SWI(+)] aggregates, colocalize only with the newly formed ring-/rod-shaped Sup35 or Rnq1 aggregates, but not with the dot-shaped mature prion aggregates. Their colocalization frequency is coordinated with their prion inducibility, indicating that prion-prion interactions mainly occur at the early initiation stage. Our results provide supportive evidence for the cross-seeding model of prionogenesis and highlight a complex interaction network among prions in yeast.
Keywords: Saccharomyces cerevisiae; [SWI+]; prion interactions; prionogenesis; protein aggregation.
Copyright © 2014 by the Genetics Society of America.
Figures





References
-
- Allen K. D., Chernova T. A., Tennant E. P., Wilkinson K. D., Chernoff Y. O., 2007. Effects of ubiquitin system alterations on the formation and loss of a yeast prion. J. Biol. Chem. 282: 3004–3013 - PubMed
-
- Bagriantsev S., Liebman S. W., 2004. Specificity of prion assembly in vivo. [PSI+] and [PIN+] form separate structures in yeast. J. Biol. Chem. 279: 51042–51048 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

