Single-molecule studies of riboswitch folding
- PMID: 24727093
- PMCID: PMC4177941
- DOI: 10.1016/j.bbagrm.2014.04.005
Single-molecule studies of riboswitch folding
Abstract
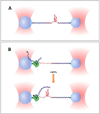
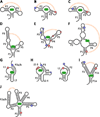
The folding dynamics of riboswitches are central to their ability to modulate gene expression in response to environmental cues. In most cases, a structural competition between the formation of a ligand-binding aptamer and an expression platform (or some other competing off-state) determines the regulatory outcome. Here, we review single-molecule studies of riboswitch folding and function, predominantly carried out using single-molecule FRET or optical trapping approaches. Recent results have supplied new insights into riboswitch folding energy landscapes, the mechanisms of ligand binding, the roles played by divalent ions, the applicability of hierarchical folding models, and kinetic vs. thermodynamic control schemes. We anticipate that future work, based on improved data sets and potentially combining multiple experimental techniques, will enable the development of more complete models for complex RNA folding processes. This article is part of a Special Issue entitled: Riboswitches.
Keywords: Gene regulation; Optical trap; Optical tweezers; Regulatory mechanism; Single molecule.
Copyright © 2014 Elsevier B.V. All rights reserved.
Figures


References
-
- Anfinsen CB. Principles that govern folding of protein chains. Science. 1973;181:223–230. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

