Ribosomal RNA gene repeats, their stability and cellular senescence
- PMID: 24727936
- PMCID: PMC4055705
- DOI: 10.2183/pjab.90.119
Ribosomal RNA gene repeats, their stability and cellular senescence
Abstract
The ribosomal RNA gene (rDNA) repeats form a historically well-researched region in the chromosome. Their highly repetitive structure can be identified easily which has enabled studies on DNA replication, recombination, and transcription. The region is one of the most unstable regions in the genome because of deleterious recombination among the repeats. The ribosomal RNA gene repeats use a unique gene amplification system to restore the copy number after this has been reduced due to recombination. It has been shown that unstable features in the genome can accelerate cellular senescence that restricts the lifespan of a cell. Here, I will introduce a study by our group that shows how the stability of rDNA is maintained and affects lifespan. I propose that the ribosomal RNA gene repeats constitute a center from which the stability of the whole genome is regulated and the lifespan of the cell is controlled.
Figures





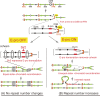


References
-
- Nomura M., Morgan E.A., Jaskunas S.R. (1977) Genetics of bacterial ribosomes. Annu. Rev. Genet. 11, 297–347 - PubMed
-
- Warner J.R. (1999) The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24, 437–440 - PubMed
-
- Hartwell, L.H. (2008) Genetics: From Genes to Genomes, 3rd Edition. McGraw-Hill, U.S.A.
-
- Brewer B.J., Fangman W.L. (1988) Mapping replication origins in yeast chromosomes. Bioessays 13, 317–322 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

