Enhanced prediction of Src homology 2 (SH2) domain binding potentials using a fluorescence polarization-derived c-Met, c-Kit, ErbB, and androgen receptor interactome
- PMID: 24728074
- PMCID: PMC4083110
- DOI: 10.1074/mcp.M113.034876
Enhanced prediction of Src homology 2 (SH2) domain binding potentials using a fluorescence polarization-derived c-Met, c-Kit, ErbB, and androgen receptor interactome
Abstract
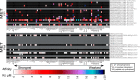
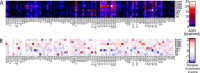
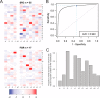
Many human diseases are associated with aberrant regulation of phosphoprotein signaling networks. Src homology 2 (SH2) domains represent the major class of protein domains in metazoans that interact with proteins phosphorylated on the amino acid residue tyrosine. Although current SH2 domain prediction algorithms perform well at predicting the sequences of phosphorylated peptides that are likely to result in the highest possible interaction affinity in the context of random peptide library screens, these algorithms do poorly at predicting the interaction potential of SH2 domains with physiologically derived protein sequences. We employed a high throughput interaction assay system to empirically determine the affinity between 93 human SH2 domains and phosphopeptides abstracted from several receptor tyrosine kinases and signaling proteins. The resulting interaction experiments revealed over 1000 novel peptide-protein interactions and provided a glimpse into the common and specific interaction potentials of c-Met, c-Kit, GAB1, and the human androgen receptor. We used these data to build a permutation-based logistic regression classifier that performed considerably better than existing algorithms for predicting the interaction potential of several SH2 domains.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



References
-
- Weinmaster G., Zoller M. J., Smith M., Hinze E., Pawson T. (1984) Mutagenesis of Fujinami sarcoma virus: evidence that tyrosine phosphorylation of P130gag-fps modulates its biological activity. Cell 37, 559–568 - PubMed
-
- Weinmaster G., Pawson T. (1986) Protein kinase activity of FSV (Fujinami sarcoma virus) P130gag-fps shows a strict specificity for tyrosine residues. J. Biol. Chem. 261, 328–333 - PubMed
-
- Anderson D., Koch C. A., Grey L., Ellis C., Moran M., F., Pawson T. (1990) Binding of SH2 domains of phospholipase C(γ)1, GAP, and Src to activated growth factor receptors. Science 250, 979–982 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

