Micro-environment causes reversible changes in DNA methylation and mRNA expression profiles in patient-derived glioma stem cells
- PMID: 24728236
- PMCID: PMC3984100
- DOI: 10.1371/journal.pone.0094045
Micro-environment causes reversible changes in DNA methylation and mRNA expression profiles in patient-derived glioma stem cells
Abstract
In vitro and in vivo models are widely used in cancer research. Characterizing the similarities and differences between a patient's tumor and corresponding in vitro and in vivo models is important for understanding the potential clinical relevance of experimental data generated with these models. Towards this aim, we analyzed the genomic aberrations, DNA methylation and transcriptome profiles of five parental tumors and their matched in vitro isolated glioma stem cell (GSC) lines and xenografts generated from these same GSCs using high-resolution platforms. We observed that the methylation and transcriptome profiles of in vitro GSCs were significantly different from their corresponding xenografts, which were actually more similar to their original parental tumors. This points to the potentially critical role of the brain microenvironment in influencing methylation and transcriptional patterns of GSCs. Consistent with this possibility, ex vivo cultured GSCs isolated from xenografts showed a tendency to return to their initial in vitro states even after a short time in culture, supporting a rapid dynamic adaptation to the in vitro microenvironment. These results show that methylation and transcriptome profiles are highly dependent on the microenvironment and growth in orthotopic sites partially reverse the changes caused by in vitro culturing.
Conflict of interest statement
Figures
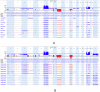
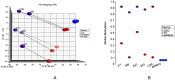
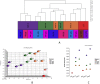
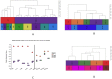
References
-
- Jones TS, Holland EC (2011) Animal models for glioma drug discovery. Expert Opinion on Drug Discovery 6: 1271–1283. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

