Whole-genome sequencing of Berkshire (European native pig) provides insights into its origin and domestication
- PMID: 24728479
- PMCID: PMC3985078
- DOI: 10.1038/srep04678
Whole-genome sequencing of Berkshire (European native pig) provides insights into its origin and domestication
Abstract
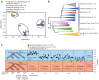
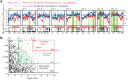
Domesticated organisms have experienced strong selective pressures directed at genes or genomic regions controlling traits of biological, agricultural or medical importance. The genome of native and domesticated pigs provide a unique opportunity for tracing the history of domestication and identifying signatures of artificial selection. Here we used whole-genome sequencing to explore the genetic relationships among the European native pig Berkshire and breeds that are distributed worldwide, and to identify genomic footprints left by selection during the domestication of Berkshire. Numerous nonsynonymous SNPs-containing genes fall into olfactory-related categories, which are part of a rapidly evolving superfamily in the mammalian genome. Phylogenetic analyses revealed a deep phylogenetic split between European and Asian pigs rather than between domestic and wild pigs. Admixture analysis exhibited higher portion of Chinese genetic material for the Berkshire pigs, which is consistent with the historical record regarding its origin. Selective sweep analyses revealed strong signatures of selection affecting genomic regions that harbor genes underlying economic traits such as disease resistance, pork yield, fertility, tameness and body length. These discoveries confirmed the history of origin of Berkshire pig by genome-wide analysis and illustrate how domestication has shaped the patterns of genetic variation.
Figures

References
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources

