Analysis of global changes in gene expression induced by human polynucleotide phosphorylase (hPNPase(old-35))
- PMID: 24729470
- PMCID: PMC4149605
- DOI: 10.1002/jcp.24645
Analysis of global changes in gene expression induced by human polynucleotide phosphorylase (hPNPase(old-35))
Abstract
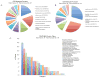
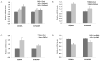
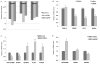
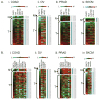
As a strategy to identify gene expression changes affected by human polynucleotide phosphorylase (hPNPase(old-35)), we performed gene expression analysis of HeLa cells in which hPNPase(old-35) was overexpressed. The observed changes were then compared to those of HO-1 melanoma cells in which hPNPase(old-35) was stably knocked down. Through this analysis, 90 transcripts, which positively or negatively correlated with hPNPase(old-35) expression, were identified. The majority of these genes were associated with cell communication, cell cycle, and chromosomal organization gene ontology categories. For a number of these genes, the positive or negative correlations with hPNPase(old-35) expression were consistent with transcriptional data extracted from the TCGA (The Cancer Genome Atlas) expression datasets for colon adenocarcinoma (COAD), skin cutaneous melanoma (SKCM), ovarian serous cyst adenocarcinoma (OV), and prostate adenocarcinoma (PRAD). Further analysis comparing the gene expression changes between Ad.hPNPase(old-35) infected HO-1 melanoma cells and HeLa cells overexpressing hPNPase(old-35) under the control of a doxycycline-inducible promoter, revealed global changes in genes involved in cell cycle and mitosis. Overall, this study provides further evidence that hPNPase(old-35) is associated with global changes in cell cycle-associated genes and identifies potential gene targets for future investigation.
© 2014 Wiley Periodicals, Inc.
Figures

References
-
- Andrade J, Pobre V, Silva I, Domingues S, Arraiano C. The role of 3′-5′ exoribonucleases in RNA degradation. Prog Mol Biol Transl Sci. 2009;85:187–229. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

