X chromosome regulation: diverse patterns in development, tissues and disease
- PMID: 24733023
- PMCID: PMC4117651
- DOI: 10.1038/nrg3687
X chromosome regulation: diverse patterns in development, tissues and disease
Abstract
Genes on the mammalian X chromosome are present in one copy in males and two copies in females. The complex mechanisms that regulate the X chromosome lead to evolutionary and physiological variability in gene expression between species, the sexes, individuals, developmental stages, tissues and cell types. In early development, delayed and incomplete X chromosome inactivation (XCI) in some species causes variability in gene expression. Additional diversity stems from escape from XCI and from mosaicism or XCI skewing in females. This causes sex-specific differences that manifest as differential gene expression and associated phenotypes. Furthermore, the complexity and diversity of X dosage regulation affect the severity of diseases caused by X-linked mutations.
Conflict of interest statement
The authors declare no competing interests.
Figures

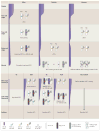
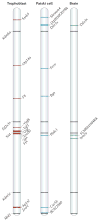
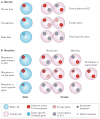
References
-
- Wang PJ, McCarrey JR, Yang F, Page DC. An abundance of X-linked genes expressed in spermatogonia. Nature Genet. 2001;27:422–426. - PubMed
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

