R PheWAS: data analysis and plotting tools for phenome-wide association studies in the R environment
- PMID: 24733291
- PMCID: PMC4133579
- DOI: 10.1093/bioinformatics/btu197
R PheWAS: data analysis and plotting tools for phenome-wide association studies in the R environment
Abstract
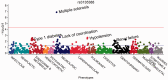
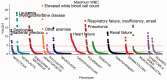
Phenome-wide association studies (PheWAS) have been used to replicate known genetic associations and discover new phenotype associations for genetic variants. This PheWAS implementation allows users to translate ICD-9 codes to PheWAS case and control groups, perform analyses using these and/or other phenotypes with covariate adjustments and plot the results. We demonstrate the methods by replicating a PheWAS on rs3135388 (near HLA-DRB, associated with multiple sclerosis) and performing a novel PheWAS using an individual's maximum white blood cell count (WBC) as a continuous measure. Our results for rs3135388 replicate known associations with more significant results than the original study on the same dataset. Our PheWAS of WBC found expected results, including associations with infections, myeloproliferative diseases and associated conditions, such as anemia. These results demonstrate the performance of the improved classification scheme and the flexibility of PheWAS encapsulated in this package.
Availability and implementation: This R package is freely available under the Gnu Public License (GPL-3) from http://phewascatalog.org. It is implemented in native R and is platform independent.
© The Author 2014. Published by Oxford University Press. All rights reserved. For Permissions, please e-mail: journals.permissions@oup.com.
Figures
References
-
- Marchini J, et al. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39:906–913. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials