Stretched cell cycle model for proliferating lymphocytes
- PMID: 24733943
- PMCID: PMC4036001
- DOI: 10.1073/pnas.1322420111
Stretched cell cycle model for proliferating lymphocytes
Abstract
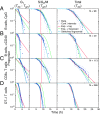
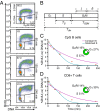
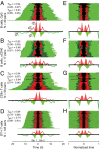
Stochastic variation in cell cycle time is a consistent feature of otherwise similar cells within a growing population. Classic studies concluded that the bulk of the variation occurs in the G1 phase, and many mathematical models assume a constant time for traversing the S/G2/M phases. By direct observation of transgenic fluorescent fusion proteins that report the onset of S phase, we establish that dividing B and T lymphocytes spend a near-fixed proportion of total division time in S/G2/M phases, and this proportion is correlated between sibling cells. This result is inconsistent with models that assume independent times for consecutive phases. Instead, we propose a stretching model for dividing lymphocytes where all parts of the cell cycle are proportional to total division time. Data fitting based on a stretched cell cycle model can significantly improve estimates of cell cycle parameters drawn from DNA labeling data used to monitor immune cell dynamics.
Keywords: FUCCI; Smith-Martin model; bromodeoxyuridine; lognormal distribution; time lapse microscopy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Howard A, Pelc SR. Nuclear incorporation of P-32 as demonstrated by autoradiographs. Exp Cell Res. 1951;2(2):178–187.
-
- Steel GG. Autoradiographic analysis of the cell cycle: Howard and Pelc to the present day. Int J Radiat Biol Relat Stud Phys Chem Med. 1986;49(2):227–235. - PubMed
-
- Killander D, Zetterberg A. Quantitative cytochemical studies on interphase growth. I. Determination of DNA, RNA and mass content of age determined mouse fibroblasts in vitro and of intercellular variation in generation time. Exp Cell Res. 1965;38(2):272–284. - PubMed
-
- Dawson KB, Madoc-Jones H, Field EO. Variations in the generation times of a strain of rat sarcoma cells in culture. Exp Cell Res. 1965;38(1):75–84. - PubMed
-
- Marin G, Bender MA. Radiation-induced mammalian cell death: Lapse-time cinemicrographic observations. Exp Cell Res. 1966;43(2):413–423. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

