Simple and cost-effective restriction endonuclease analysis of human adenoviruses
- PMID: 24734232
- PMCID: PMC3966470
- DOI: 10.1155/2014/363790
Simple and cost-effective restriction endonuclease analysis of human adenoviruses
Abstract
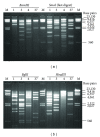
Restriction endonuclease analyses (REAs) constitute the only inexpensive molecular approach capable of typing and characterizing human adenovirus (HAdV) strains based on the entire genome. However, the application of this method is limited by the need for time-consuming and labor-intensive procedures. We herein developed a simple and cost-effective REA for assessing HAdV. The method consists of (1) simple and cost-effective DNA extraction, (2) fast restriction endonuclease (RE) digestion, and (3) speedy mini agarose gel electrophoresis. In this study, DNA was isolated according to the kit-based method and 21.0 to 28.0 μg of viral DNA was extracted from prototypes (HAdV-1, HAdV-3, HAdV-4, and HAdV-37) in each flask. The amount of DNA ranged from 11.4 to 57.0 μg among the HAdV-3 (n=73) isolates. The obtained viral DNA was found to be applicable to more than 10 types of REAs. Fast-cut restriction endonucleases (REs) were able to digest the DNA within 15 minutes, and restriction fragments were easily separated via horizontal mini agarose gel electrophoresis. The whole procedure for 10 samples can be completed within approximately six hours (the conventional method requires at least two days). These results show that our REA is potentially applicable in many laboratories in which HAdVs are isolated.
Figures

References
-
- Berk AJ. Adenoviridae. In: Knipe DM, Howley PM, Cohen JI, et al., editors. Fields Virology. 6th edition. Philadelphia, Pa, USA: Lippincott Williams & Wilkins; 2013. pp. 1704–1731.
-
- Wold WSM, Ison MG. Adenoviruses. In: Knipe DM, Howley PM, Cohen JI, et al., editors. Fields Virology. 6th edition. Philadelphia, Pa, USA: Lippincott Williams & Wilkins; 2013. pp. 1732–1767.
-
- Lion T, Kosulin K, Landlinger C, et al. Monitoring of adenovirus load in stool by real-time PCR permits early detection of impending invasive infection in patients after allogeneic stem cell transplantation. Leukemia. 2010;24(4):706–714. - PubMed
-
- Tebruegge M, Curtis N. Adenovirus infection in the immunocompromised host. Advances in Experimental Medicine and Biology. 2010;659:153–174. - PubMed
-
- Wadell G, Hammarskjold ML, Winberg G. Genetic variability of adenoviruses. Annals of the New York Academy of Sciences. 1980;354:16–42. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

