Transcriptional regulators of legume-rhizobia symbiosis: nuclear factors Ys and GRAS are two for tango
- PMID: 24736593
- PMCID: PMC4091477
- DOI: 10.4161/psb.28847
Transcriptional regulators of legume-rhizobia symbiosis: nuclear factors Ys and GRAS are two for tango
Abstract
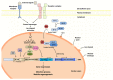
Transcription factors are DNA binding proteins that regulate gene expression. The nitrogen fixing symbiosis established between legume plants and soil bacteria is a complex interaction, in which plants need to integrate signals derived from the symbiont and the surrounding environment to initiate the developmental program of nodule organogenesis and the infection process. Several transcription factors that play critical roles in these processes have been reported in the past decade, including proteins of the GRAS and NF-Y families. Recently, we reported the characterization of a new GRAS domain containing-protein that interacts with a member of the C subunit of the NF-Y family, which plays an important role in nodule development and the progression of bacterial infection during the symbiotic interaction. The connection between transcription factors of these families highlights the significance of multimeric complexes in the fabulous capacity of plants to integrate and respond to multiple environmental stimuli.
Keywords: GRAS; lateral root; legume; nuclear factor Y; rhizobia; symbiosis.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
