Systematic identification of cell-wall related genes in Populus based on analysis of functional modules in co-expression network
- PMID: 24736620
- PMCID: PMC3988181
- DOI: 10.1371/journal.pone.0095176
Systematic identification of cell-wall related genes in Populus based on analysis of functional modules in co-expression network
Abstract
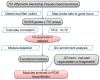
The identification of novel genes relevant to plant cell wall (PCW) biosynthesis in Populus is a highly important and challenging problem. We surveyed candidate Populus cell wall genes using a non-targeted approach. First, a genome-wide Populus gene co-expression network (PGCN) was constructed using microarray data available in the public domain. Module detection was then performed, followed by gene ontology (GO) enrichment analysis, to assign the functional category to these modules. Based on GO annotation, the modules involved in PCW biosynthesis were then selected and analyzed in detail to annotate the candidate PCW genes in these modules, including gene annotation, expression of genes in different tissues, and so on. We examined the overrepresented cis-regulatory elements (CREs) in the gene promoters to understand the possible transcriptionally co-regulated relationships among the genes within the functional modules of cell wall biosynthesis. PGCN contains 6,854 nodes (genes) with 324,238 edges. The topological properties of the network indicate scale-free and modular behavior. A total of 435 modules were identified; among which, 67 modules were identified by overrepresented GO terms. Six modules involved in cell wall biosynthesis were identified. Module 9 was mainly involved in cellular polysaccharide metabolic process in the primary cell wall, whereas Module 4 comprises genes involved in secondary cell wall biogenesis. In addition, we predicted and analyzed 10 putative CREs in the promoters of the genes in Module 4 and Module 9. The non-targeted approach of gene network analysis and the data presented here can help further identify and characterize cell wall related genes in Populus.
Conflict of interest statement
Figures

 ) in relation to the node degree for PGCN. (D) The distribution of the
) in relation to the node degree for PGCN. (D) The distribution of the  in relation to the node degree for random network.
in relation to the node degree for random network.
References
-
- Tuskan GA, DiFazio SP, Teichmann T (2004) Poplar genomics is getting popular: the impact of the poplar genome project on tree research. Plant Biol (Stuttg) 6: 2–4. - PubMed
-
- Bray D (2003) Molecular networks: the top-down view. Science 301: 1864–1865. - PubMed
-
- Jordan IK, Marino-Ramirez L, Wolf YI, Koonin EV (2004) Conservation and coevolution in the scale-free human gene coexpression network. Mol Biol Evol 21: 2058–2070. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

