Expression of a truncated ATHB17 protein in maize increases ear weight at silking
- PMID: 24736658
- PMCID: PMC3988052
- DOI: 10.1371/journal.pone.0094238
Expression of a truncated ATHB17 protein in maize increases ear weight at silking
Abstract
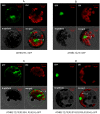
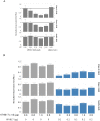
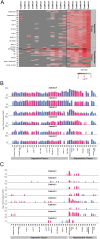
ATHB17 (AT2G01430) is an Arabidopsis gene encoding a member of the α-subclass of the homeodomain leucine zipper class II (HD-Zip II) family of transcription factors. The ATHB17 monomer contains four domains common to all class II HD-Zip proteins: a putative repression domain adjacent to a homeodomain, leucine zipper, and carboxy terminal domain. However, it also possesses a unique N-terminus not present in other members of the family. In this study we demonstrate that the unique 73 amino acid N-terminus is involved in regulation of cellular localization of ATHB17. The ATHB17 protein is shown to function as a transcriptional repressor and an EAR-like motif is identified within the putative repression domain of ATHB17. Transformation of maize with an ATHB17 expression construct leads to the expression of ATHB17Δ113, a truncated protein lacking the first 113 amino acids which encodes a significant portion of the repression domain. Because ATHB17Δ113 lacks the repression domain, the protein cannot directly affect the transcription of its target genes. ATHB17Δ113 can homodimerize, form heterodimers with maize endogenous HD-Zip II proteins, and bind to target DNA sequences; thus, ATHB17Δ113 may interfere with HD-Zip II mediated transcriptional activity via a dominant negative mechanism. We provide evidence that maize HD-Zip II proteins function as transcriptional repressors and that ATHB17Δ113 relieves this HD-Zip II mediated transcriptional repression activity. Expression of ATHB17Δ113 in maize leads to increased ear size at silking and, therefore, may enhance sink potential. We hypothesize that this phenotype could be a result of modulation of endogenous HD-Zip II pathways in maize.
Conflict of interest statement
Figures






Similar articles
-
HD-Zip proteins of families I and II from rice: interactions and functional properties.Mol Gen Genet. 2000 Feb;263(1):12-21. doi: 10.1007/pl00008671. Mol Gen Genet. 2000. PMID: 10732669
-
Systematic analysis of sequences and expression patterns of drought-responsive members of the HD-Zip gene family in maize.PLoS One. 2011;6(12):e28488. doi: 10.1371/journal.pone.0028488. Epub 2011 Dec 2. PLoS One. 2011. PMID: 22164299 Free PMC article.
-
Application of HB17, an Arabidopsis class II homeodomain-leucine zipper transcription factor, to regulate chloroplast number and photosynthetic capacity.J Exp Bot. 2013 Nov;64(14):4479-90. doi: 10.1093/jxb/ert261. Epub 2013 Sep 4. J Exp Bot. 2013. PMID: 24006420 Free PMC article.
-
Structure and function of homodomain-leucine zipper (HD-Zip) proteins.Plant Signal Behav. 2009 Feb;4(2):86-8. doi: 10.4161/psb.4.2.7692. Plant Signal Behav. 2009. PMID: 19649178 Free PMC article. Review.
-
Interplay of HD-Zip II and III transcription factors in auxin-regulated plant development.J Exp Bot. 2015 Aug;66(16):5043-53. doi: 10.1093/jxb/erv174. Epub 2015 Apr 23. J Exp Bot. 2015. PMID: 25911742 Review.
Cited by
-
The Arabidopsis transcription factor ABIG1 relays ABA signaled growth inhibition and drought induced senescence.Elife. 2016 Oct 4;5:e13768. doi: 10.7554/eLife.13768. Elife. 2016. PMID: 27697148 Free PMC article.
-
Global Analysis of Cereal microProteins Suggests Diverse Roles in Crop Development and Environmental Adaptation.G3 (Bethesda). 2020 Oct 5;10(10):3709-3717. doi: 10.1534/g3.120.400794. G3 (Bethesda). 2020. PMID: 32763954 Free PMC article.
-
Three strategies of transgenic manipulation for crop improvement.Front Plant Sci. 2022 Jul 22;13:948518. doi: 10.3389/fpls.2022.948518. eCollection 2022. Front Plant Sci. 2022. PMID: 35937379 Free PMC article. Review.
-
Assessment of genetically modified maize MON 87403 for food and feed uses, import and processing, under Regulation (EC) No 1829/2003 (application EFSA-GMO-BE-2015-125).EFSA J. 2018 Mar 28;16(3):e05225. doi: 10.2903/j.efsa.2018.5225. eCollection 2018 Mar. EFSA J. 2018. PMID: 32625854 Free PMC article.
-
Phenotypic effects from the expression of a deregulated AtGAD1 transgene and GABA pathway suppression mutants in maize.PLoS One. 2021 Dec 6;16(12):e0259365. doi: 10.1371/journal.pone.0259365. eCollection 2021. PLoS One. 2021. PMID: 34871322 Free PMC article.
References
-
- Ariel FD, Manavella PA, Dezar CA, Chan RL (2007) The true story of the HD-Zip family. Trends in Plant Science 12: 419–426. - PubMed
-
- Steindler C, Matteucci A, Sessa G, Weimar T, Ohgishi M, et al. (1999) Shade avoidance responses are mediated by the ATHB-2 HD-Zip protein, a negative regulator of gene expression. Development 126: 4235–4245. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

