Endorepellin evokes autophagy in endothelial cells
- PMID: 24737315
- PMCID: PMC4047384
- DOI: 10.1074/jbc.M114.556530
Endorepellin evokes autophagy in endothelial cells
Abstract
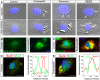
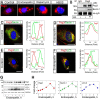
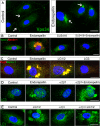
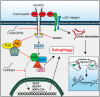
Endorepellin, the C-terminal fragment of the heparan sulfate proteoglycan perlecan, possesses angiostatic activity via dual receptor antagonism, through concurrent binding to the α2β1 integrin and vascular endothelial growth factor receptor 2 (VEGFR2). Here, we discovered that soluble endorepellin induced autophagy in endothelial cells by modulating the expression of Beclin 1, LC3, and p62, three established autophagic markers. Moreover, endorepellin evoked expression of the imprinted tumor suppressor gene Peg3 and its co-localization with Beclin 1 and LC3 in autophagosomes, suggesting a major role for this gene in endothelial cell autophagy. Mechanistically, endorepellin induced autophagy by down-regulating VEGFR2 via the two LG1/2 domains, whereas the C-terminal LG3 domain, the portion responsible for binding the α2β1 integrin, was ineffective. Endorepellin also induced transcriptional activity of the BECN1 promoter in endothelial cells, and the VEGFR2-specific tyrosine kinase inhibitor, SU5416, blocked this effect. Finally, we found a correlation between endorepellin-evoked inhibition of capillary morphogenesis and enhanced autophagy. Thus, we have identified a new role for this endogenous angiostatic fragment in inducing autophagy through a VEGFR2-dependent but α2β1 integrin-independent pathway. This novel mechanism specifically targets endothelial cells and could represent a promising new strategy to potentiate the angiostatic effect of endorepellin and perhaps other angiostatic matrix proteins.
Keywords: Angiogenesis; Autophagy; Endothelial Cell; Extracellular Matrix; Extracellular Matrix Proteins; Proteoglycan.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures





Similar articles
-
A current view of perlecan in physiology and pathology: A mosaic of functions.Matrix Biol. 2017 Jan;57-58:285-298. doi: 10.1016/j.matbio.2016.09.003. Epub 2016 Sep 6. Matrix Biol. 2017. PMID: 27613501 Free PMC article. Review.
-
Endorepellin-evoked Autophagy Contributes to Angiostasis.J Biol Chem. 2016 Sep 9;291(37):19245-56. doi: 10.1074/jbc.M116.740266. Epub 2016 Jul 19. J Biol Chem. 2016. PMID: 27435676 Free PMC article.
-
Endorepellin, the angiostatic module of perlecan, interacts with both the α2β1 integrin and vascular endothelial growth factor receptor 2 (VEGFR2): a dual receptor antagonism.J Biol Chem. 2011 Jul 22;286(29):25947-62. doi: 10.1074/jbc.M111.243626. Epub 2011 May 19. J Biol Chem. 2011. PMID: 21596751 Free PMC article.
-
Endorepellin laminin-like globular 1/2 domains bind Ig3-5 of vascular endothelial growth factor (VEGF) receptor 2 and block pro-angiogenic signaling by VEGFA in endothelial cells.FEBS J. 2013 May;280(10):2271-84. doi: 10.1111/febs.12164. Epub 2013 Feb 28. FEBS J. 2013. PMID: 23374253 Free PMC article.
-
The role of perlecan and endorepellin in the control of tumor angiogenesis and endothelial cell autophagy.Connect Tissue Res. 2015;56(5):381-91. doi: 10.3109/03008207.2015.1045297. Epub 2015 Jul 16. Connect Tissue Res. 2015. PMID: 26181327 Free PMC article. Review.
Cited by
-
Proteoglycan-driven Autophagy: A Nutrient-independent Mechanism to Control Intracellular Catabolism.J Histochem Cytochem. 2020 Nov;68(11):733-746. doi: 10.1369/0022155420937370. Epub 2020 Jul 6. J Histochem Cytochem. 2020. PMID: 32623955 Free PMC article. Review.
-
Decorin evokes reversible mitochondrial depolarization in carcinoma and vascular endothelial cells.Am J Physiol Cell Physiol. 2022 Nov 1;323(5):C1355-C1373. doi: 10.1152/ajpcell.00325.2022. Epub 2022 Aug 29. Am J Physiol Cell Physiol. 2022. PMID: 36036446 Free PMC article.
-
A current view of perlecan in physiology and pathology: A mosaic of functions.Matrix Biol. 2017 Jan;57-58:285-298. doi: 10.1016/j.matbio.2016.09.003. Epub 2016 Sep 6. Matrix Biol. 2017. PMID: 27613501 Free PMC article. Review.
-
Proteoglycan signaling in tumor angiogenesis and endothelial cell autophagy.Semin Cancer Biol. 2020 May;62:1-8. doi: 10.1016/j.semcancer.2019.05.003. Epub 2019 May 9. Semin Cancer Biol. 2020. PMID: 31078640 Free PMC article. Review.
-
Endorepellin-evoked Autophagy Contributes to Angiostasis.J Biol Chem. 2016 Sep 9;291(37):19245-56. doi: 10.1074/jbc.M116.740266. Epub 2016 Jul 19. J Biol Chem. 2016. PMID: 27435676 Free PMC article.
References
-
- Noonan D. M., Fulle A., Valente P., Cai S., Horigan E., Sasaki M., Yamada Y., Hassell J. R. (1991) The complete sequence of perlecan, a basement membrane heparan sulfate proteoglycan, reveals extensive similarity with laminin A chain, low density lipoprotein-receptor, and the neural cell adhesion molecule. J. Biol. Chem. 266, 22939–22947 - PubMed
-
- Murdoch A. D., Dodge G. R., Cohen I., Tuan R. S., Iozzo R. V. (1992) Primary structure of the human heparan sulfate proteoglycan from basement membrane (HSPG2/perlecan). A chimeric molecule with multiple domains homologous to the low density lipoprotein receptor, laminin, neural cell adhesion molecules, and epidermal growth factor. J. Biol. Chem. 267, 8544–8557 - PubMed
-
- Iozzo R. V., Pillarisetti J., Sharma B., Murdoch A. D., Danielson K. G., Uitto J., Mauviel A. (1997) Structural and functional characterization of the human perlecan gene promoter. Transcriptional activation by transforming factor-β via a nuclear factor 1-binding element. J. Biol. Chem. 272, 5219–5228 - PubMed
-
- Sharma B., Iozzo R. V. (1998) Transcriptional silencing of perlecan gene expression by interferon-γ. J. Biol. Chem. 273, 4642–4646 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

