MYC and the art of microRNA maintenance
- PMID: 24737842
- PMCID: PMC4109576
- DOI: 10.1101/cshperspect.a014175
MYC and the art of microRNA maintenance
Abstract
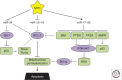
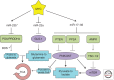
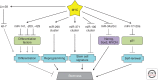
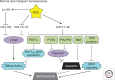
MYC is a noncanonical transcription factor that binds to thousands of genomic loci and affects >15% of the human transcriptome, with surprisingly little overlap between MYC-bound and -regulated genes. This discordance raises the question whether MYC chooses its targets based on their individual biological effects ("a la carte") or by virtue of belonging to a certain group of genes (on a "prix fixe" basis). This review presents evidence for a prix fixe, posttranscriptional model whereby MYC initially deregulates a select number of microRNAs. These microRNAs then target a broad spectrum of genes based solely on the presence in their 3' UTRs (untranslated regions) of distinct "seed" sequences. Existing evidence suggests that there are significant microRNA components to all key MYC-driven phenotypes, including cell-cycle progression, apoptosis, metabolism, angiogenesis, metastasis, stemness, and hematopoiesis. Furthermore, each of these cell-intrinsic and -extrinsic phenotypes is likely attributable to deregulation of multiple microRNA targets acting in different, yet frequently overlapping, pathways. The habitual targeting of multiple genes within the same pathway might account for the robustness and persistence of MYC-induced phenotypes.
Copyright © 2014 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures



References
-
- Abrams HD, Rohrschneider LR, Eisenman RN 1982. Nuclear location of the putative transforming protein of avian myelocytomatosis virus. Cell 29: 427–439 - PubMed
-
- Adhikary S, Eilers M 2005. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol 6: 635–645 - PubMed
-
- Bates S, Bonetta L, MacAllan D, Parry D, Holder A, Dickson C, Peters G 1994. CDK6 (PLSTIRE) and CDK4 (PSK-J3) are a distinct subset of the cyclin-dependent kinases that associate with cyclin D1. Oncogene 9: 71–79 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
