Transferable vancomycin resistance in a community-associated MRSA lineage
- PMID: 24738669
- PMCID: PMC4112484
- DOI: 10.1056/NEJMoa1303359
Transferable vancomycin resistance in a community-associated MRSA lineage
Erratum in
- N Engl J Med. 2014 Jun 5;370(23):2253
Abstract
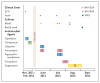
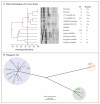
We report the case of a patient from Brazil with a bloodstream infection caused by a strain of methicillin-resistant Staphylococcus aureus (MRSA) that was susceptible to vancomycin (designated BR-VSSA) but that acquired the vanA gene cluster during antibiotic therapy and became resistant to vancomycin (designated BR-VRSA). Both strains belong to the sequence type (ST) 8 community-associated genetic lineage that carries the staphylococcal chromosomal cassette mec (SCCmec) type IVa and the S. aureus protein A gene (spa) type t292 and are phylogenetically related to MRSA lineage USA300. A conjugative plasmid of 55,706 bp (pBRZ01) carrying the vanA cluster was identified and readily transferred to other staphylococci. The pBRZ01 plasmid harbors DNA sequences that are typical of the plasmid-associated replication genes rep24 or rep21 described in community-associated MRSA strains from Australia (pWBG745). The presence and dissemination of community-associated MRSA containing vanA could become a serious public health concern.
Figures

Similar articles
-
pSK41-like plasmid is necessary for Inc18-like vanA plasmid transfer from Enterococcus faecalis to Staphylococcus aureus in vitro.Antimicrob Agents Chemother. 2013 Jan;57(1):212-9. doi: 10.1128/AAC.01587-12. Epub 2012 Oct 22. Antimicrob Agents Chemother. 2013. PMID: 23089754 Free PMC article.
-
First Report of the Local Spread of Vancomycin-Resistant Enterococci Ascribed to the Interspecies Transmission of a vanA Gene Cluster-Carrying Linear Plasmid.mSphere. 2020 Apr 8;5(2):e00102-20. doi: 10.1128/mSphere.00102-20. mSphere. 2020. PMID: 32269153 Free PMC article.
-
Characterization of SCCmec and spa types of methicillin-resistant Staphylococcus aureus isolates from health-care and community-acquired infections in Kerman, Iran.J Epidemiol Glob Health. 2017 Dec;7(4):263-267. doi: 10.1016/j.jegh.2017.08.004. Epub 2017 Aug 30. J Epidemiol Glob Health. 2017. PMID: 29110867 Free PMC article.
-
Vancomycin Resistance in Staphylococcus aureus .Yale J Biol Med. 2017 Jun 23;90(2):269-281. eCollection 2017 Jun. Yale J Biol Med. 2017. PMID: 28656013 Free PMC article. Review.
-
Broad-host-range Inc18 plasmids: Occurrence, spread and transfer mechanisms.Plasmid. 2018 Sep;99:11-21. doi: 10.1016/j.plasmid.2018.06.001. Epub 2018 Jun 19. Plasmid. 2018. PMID: 29932966 Review.
Cited by
-
Pandemic clone USA300 in a Brazilian hospital: detection of an emergent lineage among methicillin-resistant Staphylococcus aureus isolates from bloodstream infections.Antimicrob Resist Infect Control. 2022 Sep 14;11(1):114. doi: 10.1186/s13756-022-01154-3. Antimicrob Resist Infect Control. 2022. PMID: 36104710 Free PMC article.
-
Complete Genome Sequences of Eight Methicillin-Resistant Staphylococcus aureus Strains Isolated from Patients in Japan.Microbiol Resour Announc. 2019 Nov 21;8(47):e01212-19. doi: 10.1128/MRA.01212-19. Microbiol Resour Announc. 2019. PMID: 31753944 Free PMC article.
-
Antiinfective therapy with a small molecule inhibitor of Staphylococcus aureus sortase.Proc Natl Acad Sci U S A. 2014 Sep 16;111(37):13517-22. doi: 10.1073/pnas.1408601111. Epub 2014 Sep 2. Proc Natl Acad Sci U S A. 2014. PMID: 25197057 Free PMC article.
-
Molecular Characterization of Gene-Mediated Resistance and Susceptibility of ESKAPE Clinical Isolates to Cistus monspeliensis L. and Cistus salviifolius L. Extracts.Evid Based Complement Alternat Med. 2022 Sep 27;2022:7467279. doi: 10.1155/2022/7467279. eCollection 2022. Evid Based Complement Alternat Med. 2022. PMID: 36204117 Free PMC article.
-
Evolution of a 72-Kilobase Cointegrant, Conjugative Multiresistance Plasmid in Community-Associated Methicillin-Resistant Staphylococcus aureus Isolates from the Early 1990s.Antimicrob Agents Chemother. 2019 Oct 22;63(11):e01560-19. doi: 10.1128/AAC.01560-19. Print 2019 Nov. Antimicrob Agents Chemother. 2019. PMID: 31501140 Free PMC article.
References
-
- Kos VN, Desjardins CA, Griggs A, et al. Comparative genomics of vancomycin-resistant Staphylococcus aureus strains and their positions within the clade most com monly associated with methicillin-resistant S. aureus hospital-acquired infection in the United States. MBio. 2012;22:3(3):e00112–12. - PMC - PubMed
-
- Saha B, Singh AK, Ghosh A, Bal M. Identification and characterization of a vancomycin-resistant Staphylococcus aureus isolated from Kolkata (South Asia) J Med Microbiol. 2008;57:72–9. - PubMed
-
- Weigel LM, Clewell DB, Gill SR, et al. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science. 2003;302:1569–71. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
