Nucleotide regulation of the structure and dynamics of G-actin
- PMID: 24739170
- PMCID: PMC4008840
- DOI: 10.1016/j.bpj.2014.03.012
Nucleotide regulation of the structure and dynamics of G-actin
Abstract
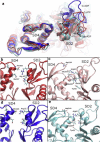
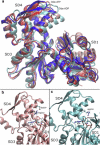
Actin, a highly conserved cytoskeletal protein found in all eukaryotic cells, facilitates cell motility and membrane remodeling via a directional polymerization cycle referred to as treadmilling. The nucleotide bound at the core of each actin subunit regulates this process. Although the biochemical kinetics of treadmilling has been well characterized, the atomistic details of how the nucleotide affects polymerization remain to be definitively determined. There is increasing evidence that the nucleotide regulation (and other characteristics) of actin cannot be fully described from the minimum energy structure, but rather depends on a dynamic equilibrium between conformations. In this work we explore the conformational mobility of the actin monomer (G-actin) in a coarse-grained subspace using umbrella sampling to bias all-atom molecular-dynamics simulations along the variables of interest. The results reveal that ADP-bound actin subunits are more conformationally mobile than ATP-bound subunits. We used a multiscale analysis method involving coarse-grained and atomistic representations of these simulations to characterize how the nucleotide affects the low-energy states of these systems. The interface between subdomains SD2-SD4, which is important for polymerization, is stabilized in an actin filament-like (F-actin) conformation in ATP-bound G-actin. Additionally, the nucleotide modulates the conformation of the SD1-SD3 interface, a region involved in the binding of several actin-binding proteins.
Copyright © 2014 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures



Similar articles
-
Nucleotide-dependence of G-actin conformation from multiple molecular dynamics simulations and observation of a putatively polymerization-competent superclosed state.Proteins. 2009 Aug 1;76(2):353-64. doi: 10.1002/prot.22350. Proteins. 2009. PMID: 19156817
-
Allostery of actin filaments: molecular dynamics simulations and coarse-grained analysis.Proc Natl Acad Sci U S A. 2005 Sep 13;102(37):13111-6. doi: 10.1073/pnas.0503732102. Epub 2005 Aug 31. Proc Natl Acad Sci U S A. 2005. PMID: 16135566 Free PMC article.
-
Nucleotide-dependent conformational states of actin.Proc Natl Acad Sci U S A. 2009 Aug 4;106(31):12723-8. doi: 10.1073/pnas.0902092106. Epub 2009 Jul 20. Proc Natl Acad Sci U S A. 2009. PMID: 19620726 Free PMC article.
-
Actin polymerization: regulation by divalent metal ion and nucleotide binding, ATP hydrolysis and binding of myosin.Adv Exp Med Biol. 1994;358:71-81. doi: 10.1007/978-1-4615-2578-3_7. Adv Exp Med Biol. 1994. PMID: 7801813 Review.
-
Actin polymerization and ATP hydrolysis.Adv Biophys. 1990;26:51-73. doi: 10.1016/0065-227x(90)90007-g. Adv Biophys. 1990. PMID: 2082729 Review.
Cited by
-
Coarse-Grained Directed Simulation.J Chem Theory Comput. 2017 Sep 12;13(9):4593-4603. doi: 10.1021/acs.jctc.7b00690. Epub 2017 Aug 31. J Chem Theory Comput. 2017. PMID: 28800392 Free PMC article.
-
Exploring Valleys without Climbing Every Peak: More Efficient and Forgiving Metabasin Metadynamics via Robust On-the-Fly Bias Domain Restriction.J Chem Theory Comput. 2015 Dec 8;11(12):5638-50. doi: 10.1021/acs.jctc.5b00907. Epub 2015 Nov 20. J Chem Theory Comput. 2015. PMID: 26587809 Free PMC article.
-
Structural basis for cofilin binding and actin filament disassembly.Nat Commun. 2018 May 10;9(1):1860. doi: 10.1038/s41467-018-04290-w. Nat Commun. 2018. PMID: 29749375 Free PMC article.
-
Histidine 73 methylation coordinates β-actin plasticity in response to key environmental factors.Nat Commun. 2025 Mar 7;16(1):2304. doi: 10.1038/s41467-025-57458-6. Nat Commun. 2025. PMID: 40055316 Free PMC article.
-
Mechanisms and therapeutic potential of disulphidptosis in cancer.Cell Prolif. 2025 Jan;58(1):e13752. doi: 10.1111/cpr.13752. Epub 2024 Oct 1. Cell Prolif. 2025. PMID: 39354653 Free PMC article. Review.
References
-
- Carlier M.F. Springer; Dordrecht, The Netherlands: 2010. Actin-Based Motility: Cellular, Molecular and Physical Aspects.
-
- Pollard T.D., Borisy G.G. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

