Deterministic processes guide long-term synchronised population dynamics in replicate anaerobic digesters
- PMID: 24739627
- PMCID: PMC4184015
- DOI: 10.1038/ismej.2014.50
Deterministic processes guide long-term synchronised population dynamics in replicate anaerobic digesters
Abstract
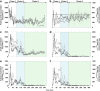
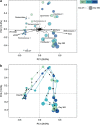
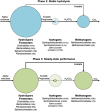
A replicate long-term experiment was conducted using anaerobic digestion (AD) as a model process to determine the relative role of niche and neutral theory on microbial community assembly, and to link community dynamics to system performance. AD is performed by a complex network of microorganisms and process stability relies entirely on the synergistic interactions between populations belonging to different functional guilds. In this study, three independent replicate anaerobic digesters were seeded with the same diverse inoculum, supplied with a model substrate, α-cellulose, and operated for 362 days at a 10-day hydraulic residence time under mesophilic conditions. Selective pressure imposed by the operational conditions and model substrate caused large reproducible changes in community composition including an overall decrease in richness in the first month of operation, followed by synchronised population dynamics that correlated with changes in reactor performance. This included the synchronised emergence and decline of distinct Ruminococcus phylotypes at day 148, and emergence of a Clostridium and Methanosaeta phylotype at day 178, when performance became stable in all reactors. These data suggest that many dynamic functional niches are predictably filled by phylogenetically coherent populations over long time scales. Neutral theory would predict that a complex community with a high degree of recognised functional redundancy would lead to stochastic changes in populations and community divergence over time. We conclude that deterministic processes may play a larger role in microbial community dynamics than currently appreciated, and under controlled conditions it may be possible to reliably predict community structural and functional changes over time.
Figures


References
-
- Albertsen M, Hugenholtz P, Skarshewski A, Nielsen KA, Tyson GW, Nielsen PH. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol. 2013;31:533–538. - PubMed
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipma DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Amani T, Nosrati M, Sreekrishnan TR. Anaerobic digestion from the viewpoint of microbiological, chemical and operational aspects - a review. Environ Rev. 2010;18:255–278.

