Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq
- PMID: 24739965
- PMCID: PMC4145853
- DOI: 10.1038/nature13173
Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq
Abstract
The mammalian lung is a highly branched network in which the distal regions of the bronchial tree transform during development into a densely packed honeycomb of alveolar air sacs that mediate gas exchange. Although this transformation has been studied by marker expression analysis and fate-mapping, the mechanisms that control the progression of lung progenitors along distinct lineages into mature alveolar cell types are still incompletely known, in part because of the limited number of lineage markers and the effects of ensemble averaging in conventional transcriptome analysis experiments on cell populations. Here we show that single-cell transcriptome analysis circumvents these problems and enables direct measurement of the various cell types and hierarchies in the developing lung. We used microfluidic single-cell RNA sequencing (RNA-seq) on 198 individual cells at four different stages encompassing alveolar differentiation to measure the transcriptional states which define the developmental and cellular hierarchy of the distal mouse lung epithelium. We empirically classified cells into distinct groups by using an unbiased genome-wide approach that did not require a priori knowledge of the underlying cell types or the previous purification of cell populations. The results confirmed the basic outlines of the classical model of epithelial cell-type diversity in the distal lung and led to the discovery of many previously unknown cell-type markers, including transcriptional regulators that discriminate between the different populations. We reconstructed the molecular steps during maturation of bipotential progenitors along both alveolar lineages and elucidated the full life cycle of the alveolar type 2 cell lineage. This single-cell genomics approach is applicable to any developing or mature tissue to robustly delineate molecularly distinct cell types, define progenitors and lineage hierarchies, and identify lineage-specific regulatory factors.
Conflict of interest statement
Figures






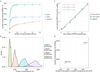

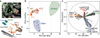
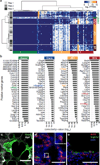
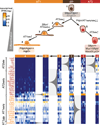

Comment in
-
Deciphering developmental processes from single-cell transcriptomes.Dev Cell. 2014 May 12;29(3):260-1. doi: 10.1016/j.devcel.2014.04.032. Dev Cell. 2014. PMID: 24823374
References
-
- Kim CFB, et al. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121:823–835. - PubMed
-
- Gonzalez R, et al. Freshly isolated rat alveolar type I cells, type II cells, and cultured type II cells have distinct molecular phenotypes. Am. J. Physiol. Lung Cell Mol. Physiol. 2005;288:L179–L189. - PubMed
Methods References
-
- Babraham Institute, Babraham Bioinformatics. FASTQC. bioinformatics.bbsrc.ac.uk. at < http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc>.
-
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. 2011. 2011;17
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

