Focused specificity of intestinal TH17 cells towards commensal bacterial antigens
- PMID: 24739972
- PMCID: PMC4128479
- DOI: 10.1038/nature13279
Focused specificity of intestinal TH17 cells towards commensal bacterial antigens
Abstract
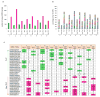
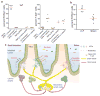
T-helper-17 (TH17) cells have critical roles in mucosal defence and in autoimmune disease pathogenesis. They are most abundant in the small intestine lamina propria, where their presence requires colonization of mice with microbiota. Segmented filamentous bacteria (SFB) are sufficient to induce TH17 cells and to promote TH17-dependent autoimmune disease in animal models. However, the specificity of TH17 cells, the mechanism of their induction by distinct bacteria, and the means by which they foster tissue-specific inflammation remain unknown. Here we show that the T-cell antigen receptor (TCR) repertoire of intestinal TH17 cells in SFB-colonized mice has minimal overlap with that of other intestinal CD4(+) T cells and that most TH17 cells, but not other T cells, recognize antigens encoded by SFB. T cells with antigen receptors specific for SFB-encoded peptides differentiated into RORγt-expressing TH17 cells, even if SFB-colonized mice also harboured a strong TH1 cell inducer, Listeria monocytogenes, in their intestine. The match of T-cell effector function with antigen specificity is thus determined by the type of bacteria that produce the antigen. These findings have significant implications for understanding how commensal microbiota contribute to organ-specific autoimmunity and for developing novel mucosal vaccines.
Figures

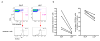










References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

