Whole CMV proteome pattern recognition analysis after HSCT identifies unique epitope targets associated with the CMV status
- PMID: 24740411
- PMCID: PMC3989190
- DOI: 10.1371/journal.pone.0089648
Whole CMV proteome pattern recognition analysis after HSCT identifies unique epitope targets associated with the CMV status
Abstract
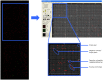
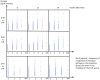
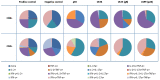
Cytomegalovirus (CMV) infection represents a vital complication after Hematopoietic Stem Cell Transplantation (HSCT). We screened the entire CMV proteome to visualize the humoral target epitope-focus profile in serum after HSCT. IgG profiling from four patient groups (donor and/or recipient +/- for CMV) was performed at 6, 12 and 24 months after HSCT using microarray slides containing 17174 of 15mer-peptides overlapping by 4 aa covering 214 proteins from CMV. Data were analyzed using maSigPro, PAM and the 'exclusive recognition analysis (ERA)' to identify unique CMV epitope responses for each patient group. The 'exclusive recognition analysis' of serum epitope patterns segregated best 12 months after HSCT for the D+/R+ group (versus D-/R-). Epitopes were derived from UL123 (IE1), UL99 (pp28), UL32 (pp150), this changed at 24 months to 2 strongly recognized peptides provided from UL123 and UL100. Strongly (IgG) recognized CMV targets elicited also robust cytokine production in T-cells from patients after HSCT defined by intracellular cytokine staining (IL-2, TNF, IFN and IL-17). High-content peptide microarrays allow epitope profiling of entire viral proteomes; this approach can be useful to map relevant targets for diagnostics and therapy in patients with well defined clinical endpoints. Peptide microarray analysis visualizes the breadth of B-cell immune reconstitution after HSCT and provides a useful tool to gauge immune reconstitution.
Conflict of interest statement
Figures


References
-
- Ljungman P, Perez-Bercoff L, Jonsson J, Avetisyan G, Sparrelid E, et al. (2006) Risk factors for the development of cytomegalovirus disease after allogeneic stem cell transplantation. Haematologica 91: 78–83. - PubMed
-
- Weseslindtner L, Kerschner H, Steinacher D, Nachbagauer R, Kundi M, et al. (2012) Prospective Analysis of Human Cytomegalovirus DNAemia and Specific CD8+ T Cell Responses in Lung Transplant Recipients. Am J Transplant 12: 2172–2180. - PubMed
-
- Schoppel K, Schmidt C, Einsele H, Hebart H, Mach M (1998) Kinetics of the antibody response against human cytomegalovirus-specific proteins in allogeneic bone marrow transplant recipients. J Infect Dis 178: 1233–1243. - PubMed
-
- Ruutu T, Ljungman P, Brinch L, Lenhoff S, Lonnqvist B, et al. (1997) No prevention of cytomegalovirus infection by anti-cytomegalovirus hyperimmune globulin in seronegative bone marrow transplant recipients. The Nordic BMT Group. Bone Marrow Transplantation 19: 233–236. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

