Bridging the gap between the human and macaque connectome: a quantitative comparison of global interspecies structure-function relationships and network topology
- PMID: 24741045
- PMCID: PMC3988411
- DOI: 10.1523/JNEUROSCI.4229-13.2014
Bridging the gap between the human and macaque connectome: a quantitative comparison of global interspecies structure-function relationships and network topology
Abstract
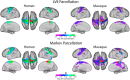
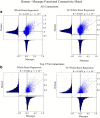
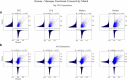
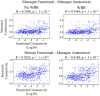
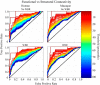
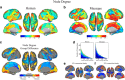
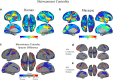
Resting state functional connectivity MRI (rs-fcMRI) may provide a powerful and noninvasive "bridge" for comparing brain function between patients and experimental animal models; however, the relationship between human and macaque rs-fcMRI remains poorly understood. Here, using a novel surface deformation process for species comparisons in the same anatomical space (Van Essen, 2004, 2005), we found high correspondence, but also unique hub topology, between human and macaque functional connectomes. The global functional connectivity match between species was moderate to strong (r = 0.41) and increased when considering the top 15% strongest connections (r = 0.54). Analysis of the match between functional connectivity and the underlying anatomical connectivity, derived from a previous retrograde tracer study done in macaques (Markov et al., 2012), showed impressive structure-function correspondence in both the macaque and human. When examining the strongest structural connections, we found a 70-80% match between structural and functional connectivity matrices in both species. Finally, we compare species on two widely used metrics for studying hub topology: degree and betweenness centrality. The data showed topological agreement across the species, with nodes of the posterior cingulate showing high degree and betweenness centrality. In contrast, nodes in medial frontal and parietal cortices were identified as having high degree and betweenness in the human as opposed to the macaque. Our results provide: (1) a thorough examination and validation for a surface-based interspecies deformation process, (2) a strong theoretical foundation for making interspecies comparisons of rs-fcMRI, and (3) a unique look at topological distinctions between the species.
Keywords: graph theory; macaque functional connectivity; network topology; resting state functional connectivity MRI; structure function relationships.
Figures



References
-
- Biederman J, Faraone SV, Keenan K, Benjamin J, Krifcher B, Moore C, Sprich-Buckminster S, Ugaglia K, Jellinek MS, Steingard R. Further evidence for family-genetic risk factors in attention deficit hyperactivity disorder: patterns of comorbidity in probands and relatives in psychiatrically and pediatrically referred samples. Arch Gen Psychiatry. 1992;49:728–738. doi: 10.1001/archpsyc.1992.01820090056010. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
