Characteristics of nucleocytoplasmic transport of H1N1 influenza A virus nuclear export protein
- PMID: 24741105
- PMCID: PMC4054460
- DOI: 10.1128/JVI.00257-14
Characteristics of nucleocytoplasmic transport of H1N1 influenza A virus nuclear export protein
Abstract
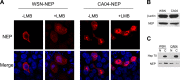
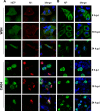
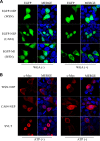
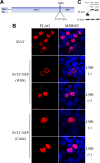
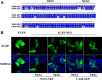
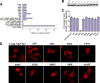
The influenza A virus nuclear export protein (NEP) plays crucial roles in the nuclear export of the viral ribonucleoprotein complex through the chromosome region maintenance 1 (CRM1)-mediated cellular protein transport system. However, the detailed mechanism of NEP nucleocytoplasmic trafficking remains incompletely understood. Here, we investigated the subcellular localization of NEP from two strains of H1N1 influenza A virus and found that 2009 swine-origin H1N1 influenza A virus A/California/04/2009 (CA04) NEP displayed a distinct cellular distribution pattern, forming unique nuclear aggregates, compared to A/WSN/33 (H1N1) (WSN) NEP. Characterization of the nucleocytoplasmic transport pathways of these two NEPs showed that they both enter the nucleus by passive diffusion but are exported through the nuclear export receptor CRM1-mediated pathway with different efficiencies. The two identified nuclear export signals (NESs) on the two NEPs functioned similarly despite differences in their amino acid sequences. Using a two-hybrid assay, we confirmed that the CA04 NEP interacts less efficiently with CRM1 and that a threonine residue at position 48 is responsible for the nuclear aggregation. The present study revealed the dissimilarity in subcellular NEP transport processes between the 2009 pandemic (H1N1) influenza A virus CA04 and the laboratory-adapted H1N1 virus WSN and uncovered the mechanism responsible for this difference.
Importance: Because the efficiency of the nucleocytoplasmic transport of viral components is often correlated with the viral RNA polymerase activity, propagation, and host range of influenza viruses, the present study investigated the subcellular localization of NEP from two strains of H1N1 influenza virus. We found that the NEPs of both A/California/04/2009 (H1N1) (CA04) and A/WSN/33 (H1N1) (WSN) enter the nucleus by passive diffusion but are exported with different efficiencies, which were caused by weaker binding activity between the CA04 NEP and CRM1. The results of the present study revealed characteristics of the nuclear import and export pathways of NEP and the mechanism responsible for the differences in the cellular distribution of NEP between two H1N1 strains.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures
Similar articles
-
Nucleocytoplasmic Shuttling of Porcine Parvovirus NS1 Protein Mediated by the CRM1 Nuclear Export Pathway and the Importin α/β Nuclear Import Pathway.J Virol. 2022 Jan 12;96(1):e0148121. doi: 10.1128/JVI.01481-21. Epub 2021 Oct 13. J Virol. 2022. PMID: 34643426 Free PMC article.
-
NXT1, a Novel Influenza A NP Binding Protein, Promotes the Nuclear Export of NP via a CRM1-Dependent Pathway.Viruses. 2016 Jul 28;8(8):209. doi: 10.3390/v8080209. Viruses. 2016. PMID: 27483302 Free PMC article.
-
Differential nucleocytoplasmic shuttling of the nucleoprotein of influenza a viruses and association with host tropism.Cell Microbiol. 2017 May;19(5). doi: 10.1111/cmi.12692. Epub 2016 Nov 25. Cell Microbiol. 2017. PMID: 27862840
-
Strength in Diversity: Nuclear Export of Viral RNAs.Viruses. 2020 Sep 11;12(9):1014. doi: 10.3390/v12091014. Viruses. 2020. PMID: 32932882 Free PMC article. Review.
-
Viral Subversion of the Chromosome Region Maintenance 1 Export Pathway and Its Consequences for the Cell Host.Viruses. 2023 Nov 6;15(11):2218. doi: 10.3390/v15112218. Viruses. 2023. PMID: 38005895 Free PMC article. Review.
Cited by
-
Phosphorylation of S-S-S Motif in Nuclear Export Protein (NEP) Plays a Critical Role in Viral Ribonucleoprotein (vRNP) Nuclear Export of Influenza A and B Viruses.Adv Sci (Weinh). 2025 Jan;12(2):e2309477. doi: 10.1002/advs.202309477. Epub 2024 Nov 22. Adv Sci (Weinh). 2025. PMID: 39575547 Free PMC article.
-
How SARS-CoV-2 and Other Viruses Build an Invasion Route to Hijack the Host Nucleocytoplasmic Trafficking System.Cells. 2021 Jun 7;10(6):1424. doi: 10.3390/cells10061424. Cells. 2021. PMID: 34200500 Free PMC article. Review.
-
Nucleocytoplasmic shuttling of influenza A virus proteins.Viruses. 2015 May 22;7(5):2668-82. doi: 10.3390/v7052668. Viruses. 2015. PMID: 26008706 Free PMC article. Review.
-
PA and PA-X: two key proteins from segment 3 of the influenza viruses.Front Cell Infect Microbiol. 2025 Mar 14;15:1560250. doi: 10.3389/fcimb.2025.1560250. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40160474 Free PMC article. Review.
-
Investigating the Role of African Horse Sickness Virus VP7 Protein Crystalline Particles on Virus Replication and Release.Viruses. 2022 Oct 4;14(10):2193. doi: 10.3390/v14102193. Viruses. 2022. PMID: 36298748 Free PMC article.
References
-
- Jagger BW, Wise HM, Kash JC, Walters KA, Wills NM, Xiao YL, Dunfee RL, Schwartzman LM, Ozinsky A, Bell GL, Dalton RM, Lo A, Efstathiou S, Atkins JF, Firth AE, Taubenberger JK, Digard P. 2012. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science 337:199–204. 10.1126/science.1222213 - DOI - PMC - PubMed
-
- Wise HM, Hutchinson EC, Jagger BW, Stuart AD, Kang ZH, Robb N, Schwartzman LM, Kash JC, Fodor E, Firth AE, Gog JR, Taubenberger JK, Digard P. 2012. Identification of a novel splice variant form of the influenza A virus M2 ion channel with an antigenically distinct ectodomain. PLoS Pathog. 8:e1002998. 10.1371/journal.ppat.1002998 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

