NS2 proteins of GB virus B and hepatitis C virus share common protease activities and membrane topologies
- PMID: 24741107
- PMCID: PMC4054453
- DOI: 10.1128/JVI.00656-14
NS2 proteins of GB virus B and hepatitis C virus share common protease activities and membrane topologies
Abstract
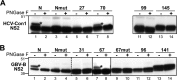
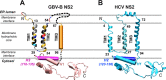
GB virus B (GBV-B), which is hepatotropic in experimentally infected small New World primates, is a member of the Hepacivirus genus but phylogenetically relatively distant from hepatitis C virus (HCV). To gain insights into the role and specificity of hepaciviral nonstructural protein 2 (NS2), which is required for HCV polyprotein processing and particle morphogenesis, we investigated whether NS2 structural and functional features are conserved between HCV and GBV-B. We found that GBV-B NS2, like HCV NS2, has cysteine protease activity responsible for cleavage at the NS2/NS3 junction, and we experimentally confirmed the location of this junction within the viral polyprotein. A model for GBV-B NS2 membrane topology was experimentally established by determining the membrane association properties of NS2 segments fused to green fluorescent protein (GFP) and their nuclear magnetic resonance structures using synthetic peptides as well as by applying an N-glycosylation scanning approach. Similar glycosylation studies confirmed the HCV NS2 organization. Together, our data show that despite limited amino acid sequence similarity, GBV-B and HCV NS2 proteins share a membrane topology with 3 N-terminal transmembrane segments, which is also predicted to apply to other recently discovered hepaciviruses. Based on these data and using trans-complementation systems, we found that intragenotypic hybrid NS2 proteins with heterologous N-terminal membrane segments were able to efficiently trans-complement an assembly-deficient HCV mutant with a point mutation in the NS2 C-terminal domain, while GBV-B/HCV or intergenotypic NS2 chimeras were not. These studies indicate that virus- and genotype-specific intramolecular interactions between N- and C-terminal domains of NS2 are critically involved in HCV morphogenesis.
Importance: Nonstructural protein 2 (NS2) of hepatitis C virus (HCV) is a multifunctional protein critically involved in polyprotein processing and virion morphogenesis. To gain insights into NS2 mechanisms of action, we investigated whether NS2 structural and functional features are conserved between HCV and GB virus B (GBV-B), a phylogenetically relatively distant primate hepacivirus. We showed that GBV-B NS2, like HCV NS2, carries cysteine protease activity. We experimentally established a model for GBV-B NS2 membrane topology and demonstrated that despite limited sequence similarity, GBV-B and HCV NS2 share an organization with three N-terminal transmembrane segments. We found that the role of HCV NS2 in particle assembly is genotype specific and relies on critical interactions between its N- and C-terminal domains. This first comparative analysis of NS2 proteins from two hepaciviruses and our structural predictions of NS2 from other newly identified mammal hepaciviruses highlight conserved key features of the hepaciviral life cycle.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures








Similar articles
-
Infection of Common Marmosets with GB Virus B Chimeric Virus Encoding the Major Nonstructural Proteins NS2 to NS4A of Hepatitis C Virus.J Virol. 2016 Aug 26;90(18):8198-211. doi: 10.1128/JVI.02653-15. Print 2016 Sep 15. J Virol. 2016. PMID: 27384651 Free PMC article.
-
Determinants Involved in Hepatitis C Virus and GB Virus B Primate Host Restriction.J Virol. 2015 Dec;89(23):12131-44. doi: 10.1128/JVI.01161-15. Epub 2015 Sep 23. J Virol. 2015. PMID: 26401036 Free PMC article.
-
NS2 proteases from hepatitis C virus and related hepaciviruses share composite active sites and previously unrecognized intrinsic proteolytic activities.PLoS Pathog. 2018 Feb 7;14(2):e1006863. doi: 10.1371/journal.ppat.1006863. eCollection 2018 Feb. PLoS Pathog. 2018. PMID: 29415072 Free PMC article.
-
GB virus B as a model for hepatitis C virus.ILAR J. 2001;42(2):152-60. doi: 10.1093/ilar.42.2.152. ILAR J. 2001. PMID: 11406717 Review.
-
The hepatitis C virus NS2/3 protease.Curr Issues Mol Biol. 2007 Jan;9(1):63-9. Curr Issues Mol Biol. 2007. PMID: 17263146 Review.
Cited by
-
Infection of Common Marmosets with GB Virus B Chimeric Virus Encoding the Major Nonstructural Proteins NS2 to NS4A of Hepatitis C Virus.J Virol. 2016 Aug 26;90(18):8198-211. doi: 10.1128/JVI.02653-15. Print 2016 Sep 15. J Virol. 2016. PMID: 27384651 Free PMC article.
-
Differential regulation of the Wnt/β-catenin pathway by hepatitis C virus recombinants expressing core from various genotypes.Sci Rep. 2018 Jul 25;8(1):11185. doi: 10.1038/s41598-018-29078-2. Sci Rep. 2018. PMID: 30046100 Free PMC article.
-
Modeling HCV disease in animals: virology, immunology and pathogenesis of HCV and GBV-B infections.Front Microbiol. 2014 Dec 8;5:690. doi: 10.3389/fmicb.2014.00690. eCollection 2014. Front Microbiol. 2014. PMID: 25538700 Free PMC article. Review.
-
Identification of NS2 determinants stimulating intrinsic HCV NS2 protease activity.PLoS Pathog. 2022 Jun 21;18(6):e1010644. doi: 10.1371/journal.ppat.1010644. eCollection 2022 Jun. PLoS Pathog. 2022. PMID: 35727826 Free PMC article.
-
Determinants Involved in Hepatitis C Virus and GB Virus B Primate Host Restriction.J Virol. 2015 Dec;89(23):12131-44. doi: 10.1128/JVI.01161-15. Epub 2015 Sep 23. J Virol. 2015. PMID: 26401036 Free PMC article.
References
-
- Simons JN, Pilot-Matias TJ, Leary TP, Dawson GJ, Desai SM, Schlauder GG, Muerhoff AS, Erker JC, Buijk SL, Chalmers ML, Van Sant CL, Mushahwar IK. 1995. Identification of two flavivirus-like genomes in the GB hepatitis agent. Proc. Natl. Acad. Sci. U. S. A. 92:3401–3405. 10.1073/pnas.92.8.3401 - DOI - PMC - PubMed
-
- Iwasaki Y, Mori K, Ishii K, Maki N, Iijima S, Yoshida T, Okabayashi S, Katakai Y, Lee YJ, Saito A, Fukai H, Kimura N, Ageyama N, Yoshizaki S, Suzuki T, Yasutomi Y, Miyamura T, Kannagi M, Akari H. 2011. Long-term persistent GBV-B infection and development of a chronic and progressive hepatitis C-like disease in marmosets. Front. Microbiol. 2:240. 10.3389/fmicb.2011.00240 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

