Investigation of the genetic diversity of domestic Capra hircus breeds reared within an early goat domestication area in Iran
- PMID: 24742145
- PMCID: PMC4044659
- DOI: 10.1186/1297-9686-46-27
Investigation of the genetic diversity of domestic Capra hircus breeds reared within an early goat domestication area in Iran
Abstract
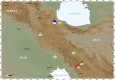
Background: Iran is an area of particular interest for investigating goat diversity. Archaeological remains indicate early goat domestication (about 10,000 years ago) in the Iranian Zagros Mountains as well as in the high Euphrates valley and southeastern Anatolia. In addition, mitochondrial DNA data of domestic goats and wild ancestors (C. aegagrusor bezoar) suggest a pre-domestication management of wild populations in southern Zagros and central Iranian Plateau. In this study genetic diversity was assessed in seven Iranian native goat breeds, namely Markhoz, Najdi, Taleshi, Khalkhali, Naini, native Abadeh and Turki-Ghashghaei. A total of 317 animals were characterized using 14 microsatellite loci. Two Pakistani goat populations, Pahari and Teddy, were genotyped for comparison.
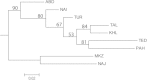
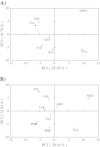
Results: Iranian goats possess a remarkable genetic diversity (average expected heterozygosity of 0.671 across loci, 10.7 alleles per locus) mainly accounted for by the within-breed component (GST = 5.9%). Positive and highly significant FIS values in the Naini, Turki-Ghashghaei, Abadeh and Markhoz breeds indicate some level of inbreeding in these populations. Multivariate analyses cluster Iranian goats into northern, central and western groups, with the western breeds relatively distinct from the others. Pakistani breeds show some relationship with Iranian populations, even if their position is not consistent across analyses. Gene flow was higher within regions (west, north, central) compared to between regions but particularly low between the western and the other two regions, probably due to the isolating topography of the Zagros mountain range. The Turki-Ghashghaei, Najdi and Abadeh breeds are reared in geographic areas where mtDNA provided evidence of early domestication. These breeds are highly variable, located on basal short branches in the neighbor-joining tree, close to the origin of the principal component analysis plot and, although highly admixed, they are quite distinct from those reared on the western side of the Zagros mountain range.
Conclusions: These observations call for further investigation of the nuclear DNA diversity of these breeds within a much wider geographic context to confirm or re-discuss the current hypothesis (based on maternal lineage data) of an almost exclusive contribution of the eastern Anatolian bezoar to the domestic goat gene pool.
Figures

Similar articles
-
The goat domestication process inferred from large-scale mitochondrial DNA analysis of wild and domestic individuals.Proc Natl Acad Sci U S A. 2008 Nov 18;105(46):17659-64. doi: 10.1073/pnas.0804782105. Epub 2008 Nov 12. Proc Natl Acad Sci U S A. 2008. PMID: 19004765 Free PMC article.
-
Mitochondrial DNA diversity of Anatolian indigenous domestic goats.J Anim Breed Genet. 2014 Dec;131(6):487-95. doi: 10.1111/jbg.12096. Epub 2014 Jun 18. J Anim Breed Genet. 2014. PMID: 24942987
-
Investigating the footprint of post-domestication dispersal on the diversity of modern European, African and Asian goats.Genet Sel Evol. 2024 Jul 27;56(1):55. doi: 10.1186/s12711-024-00923-5. Genet Sel Evol. 2024. PMID: 39068382 Free PMC article.
-
Multilocus genotypic data reveal high genetic diversity and low population genetic structure of Iranian indigenous sheep.Anim Genet. 2016 Aug;47(4):463-70. doi: 10.1111/age.12429. Epub 2016 Mar 8. Anim Genet. 2016. PMID: 26953226
-
A comprehensive review of HVS-I mitochondrial DNA variation of 19 Iranian populations.Ann Hum Genet. 2024 May;88(3):259-277. doi: 10.1111/ahg.12544. Epub 2023 Dec 31. Ann Hum Genet. 2024. PMID: 38161274 Review.
Cited by
-
Characteristics of the Allele Pool and the Genetic Differentiation of Goats of Different Breeds and their Wild Relatives by Str-Markers.Arch Razi Inst. 2021 Nov 30;76(5):1351-1362. doi: 10.22092/ari.2021.355684.1709. eCollection 2021 Nov. Arch Razi Inst. 2021. PMID: 35355766 Free PMC article.
-
Indigenous cattle of Sri Lanka: Genetic and phylogeographic relationship with Zebu of Indus Valley and South Indian origin.PLoS One. 2023 Aug 16;18(8):e0282761. doi: 10.1371/journal.pone.0282761. eCollection 2023. PLoS One. 2023. PMID: 37585485 Free PMC article.
-
Analysis of whole-genome re-sequencing data of ducks reveals a diverse demographic history and extensive gene flow between Southeast/South Asian and Chinese populations.Genet Sel Evol. 2021 Apr 13;53(1):35. doi: 10.1186/s12711-021-00627-0. Genet Sel Evol. 2021. PMID: 33849442 Free PMC article.
-
Genome-wide SNP profiling of worldwide goat populations reveals strong partitioning of diversity and highlights post-domestication migration routes.Genet Sel Evol. 2018 Nov 19;50(1):58. doi: 10.1186/s12711-018-0422-x. Genet Sel Evol. 2018. PMID: 30449284 Free PMC article.
-
Polymorphisms and mRNA Expression Levels of IGF-1, FGF5, and KAP 1.4 in Tibetan Cashmere Goats.Genes (Basel). 2023 Mar 14;14(3):711. doi: 10.3390/genes14030711. Genes (Basel). 2023. PMID: 36980983 Free PMC article.
References
-
- Tavakolian J. An Introduction to Genetic Resources of Native Farm Animals in Iran. Tehran: Animal Science Genetic Research Institute Press; 2000.
-
- Kumar S, Dixit SP, Verma NK, Singh DK, Pande A, Kumar S, Chander R, Singh LB. Genetic diversity analysis of the Gohilwari breed of Indian goat (Capra hircus) using microsatellite markers. Am J Anim Vet Sci. 2009;4:49–57.
-
- Fan B, Han JL, Chen SL, Mburu DN, Hanotte O, Chen QK, Zhao SH, Li K. Individual-breed assignments in caprine populations using microsatellite DNA analysis. Small Ruminant Res. 2008;75:154–161. doi: 10.1016/j.smallrumres.2007.09.007. - DOI
-
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor: Cold Spring Harbor Press; 1989.
-
- FAO. Molecular genetic characterization of animal genetic resources. FAO Animal Production and Health Guidelines. 2011. p. 9. http://www.fao.org/docrep/014/i2413e/i2413e00.pdf.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

