A comprehensive study of small non-frameshift insertions/deletions in proteins and prediction of their phenotypic effects by a machine learning method (KD4i)
- PMID: 24742296
- PMCID: PMC4021375
- DOI: 10.1186/1471-2105-15-111
A comprehensive study of small non-frameshift insertions/deletions in proteins and prediction of their phenotypic effects by a machine learning method (KD4i)
Abstract
Background: Small insertion and deletion polymorphisms (Indels) are the second most common mutations in the human genome, after Single Nucleotide Polymorphisms (SNPs). Recent studies have shown that they have significant influence on genetic variation by altering human traits and can cause multiple human diseases. In particular, many Indels that occur in protein coding regions are known to impact the structure or function of the protein. A major challenge is to predict the effects of these Indels and to distinguish between deleterious and neutral variants. When an Indel occurs within a coding region, it can be either frameshifting (FS) or non-frameshifting (NFS). FS-Indels either modify the complete C-terminal region of the protein or result in premature termination of translation. NFS-Indels insert/delete multiples of three nucleotides leading to the insertion/deletion of one or more amino acids.
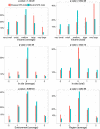
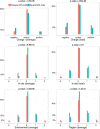
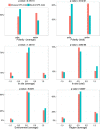
Results: In order to study the relationships between NFS-Indels and Mendelian diseases, we characterized NFS-Indels according to numerous structural, functional and evolutionary parameters. We then used these parameters to identify specific characteristics of disease-causing and neutral NFS-Indels. Finally, we developed a new machine learning approach, KD4i, that can be used to predict the phenotypic effects of NFS-Indels.
Conclusions: We demonstrate in a large-scale evaluation that the accuracy of KD4i is comparable to existing state-of-the-art methods. However, a major advantage of our approach is that we also provide the reasons for the predictions, in the form of a set of rules. The rules are interpretable by non-expert humans and they thus represent new knowledge about the relationships between the genotype and phenotypes of NFS-Indels and the causative molecular perturbations that result in the disease.
Figures








References
-
- Carling T, Correa P, Hessman O, Hedberg J, Skogseid B, Lindberg D, Rastad J, Westin G, Akerstrom G. Parathyroid MEN1 gene mutations in relation to clinical characteristics of nonfamilial primary hyperparathyroidism. J Clin Endocrinol Metab. 1998;83(8):2960–2963. - PubMed
-
- Collins FS, Brooks LD, Chakravarti A. A DNA polymorphism discovery resource for research on human genetic variation. Genome Res. 1998;8(12):1229–1231. - PubMed
-
- Friedrich A, Garnier N, Gagniere N, Nguyen H, Albou LP, Biancalana V, Bettler E, Deleage G, Lecompte O, Muller J, Moras D, Mandel JL, Toursel T, Moulinier L, Poch O. SM2PH-db: an interactive system for the integrated analysis of phenotypic consequences of missense mutations in proteins involved in human genetic diseases. Hum Mutat. 2010;31(2):127–135. doi: 10.1002/humu.21155. - DOI - PubMed
-
- Hunter DJ. Gene-environment interactions in human diseases. Nat Rev Genet. 2005;6(4):287–298. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

