Genome-wide profiling of yeast DNA:RNA hybrid prone sites with DRIP-chip
- PMID: 24743342
- PMCID: PMC3990523
- DOI: 10.1371/journal.pgen.1004288
Genome-wide profiling of yeast DNA:RNA hybrid prone sites with DRIP-chip
Abstract
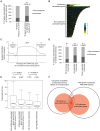
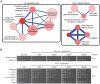
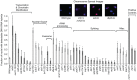
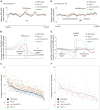
DNA:RNA hybrid formation is emerging as a significant cause of genome instability in biological systems ranging from bacteria to mammals. Here we describe the genome-wide distribution of DNA:RNA hybrid prone loci in Saccharomyces cerevisiae by DNA:RNA immunoprecipitation (DRIP) followed by hybridization on tiling microarray. These profiles show that DNA:RNA hybrids preferentially accumulated at rDNA, Ty1 and Ty2 transposons, telomeric repeat regions and a subset of open reading frames (ORFs). The latter are generally highly transcribed and have high GC content. Interestingly, significant DNA:RNA hybrid enrichment was also detected at genes associated with antisense transcripts. The expression of antisense-associated genes was also significantly altered upon overexpression of RNase H, which degrades the RNA in hybrids. Finally, we uncover mutant-specific differences in the DRIP profiles of a Sen1 helicase mutant, RNase H deletion mutant and Hpr1 THO complex mutant compared to wild type, suggesting different roles for these proteins in DNA:RNA hybrid biology. Our profiles of DNA:RNA hybrid prone loci provide a resource for understanding the properties of hybrid-forming regions in vivo, extend our knowledge of hybrid-mitigating enzymes, and contribute to models of antisense-mediated gene regulation. A summary of this paper was presented at the 26th International Conference on Yeast Genetics and Molecular Biology, August 2013.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

