A draft genome of the honey bee trypanosomatid parasite Crithidia mellificae
- PMID: 24743507
- PMCID: PMC3990616
- DOI: 10.1371/journal.pone.0095057
A draft genome of the honey bee trypanosomatid parasite Crithidia mellificae
Abstract
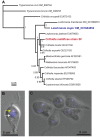
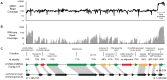
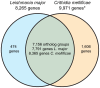
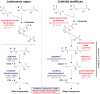
Since 2006, honey bee colonies in North America and Europe have experienced increased annual mortality. These losses correlate with increased pathogen incidence and abundance, though no single etiologic agent has been identified. Crithidia mellificae is a unicellular eukaryotic honey bee parasite that has been associated with colony losses in the USA and Belgium. C. mellificae is a member of the family Trypanosomatidae, which primarily includes other insect-infecting species (e.g., the bumble bee pathogen Crithidia bombi), as well as species that infect both invertebrate and vertebrate hosts including human pathogens (e.g.,Trypanosoma cruzi, T. brucei, and Leishmania spp.). To better characterize C. mellificae, we sequenced the genome and transcriptome of strain SF, which was isolated and cultured in 2010. The 32 megabase draft genome, presented herein, shares a high degree of conservation with the related species Leishmania major. We estimate that C. mellificae encodes over 8,300 genes, the majority of which are orthologs of genes encoded by L. major and other Leishmania or Trypanosoma species. Genes unique to C. mellificae, including those of possible bacterial origin, were annotated based on function and include genes putatively involved in carbohydrate metabolism. This draft genome will facilitate additional investigations of the impact of C. mellificae infection on honey bee health and provide insight into the evolution of this unique family.
Conflict of interest statement
Figures
Similar articles
-
Differential diagnosis of the honey bee trypanosomatids Crithidia mellificae and Lotmaria passim.J Invertebr Pathol. 2015 Sep;130:21-7. doi: 10.1016/j.jip.2015.06.007. Epub 2015 Jul 3. J Invertebr Pathol. 2015. PMID: 26146231
-
Double-stranded RNA reduces growth rates of the gut parasite Crithidia mellificae.Parasitol Res. 2019 Feb;118(2):715-721. doi: 10.1007/s00436-018-6176-0. Epub 2019 Jan 4. Parasitol Res. 2019. PMID: 30607610
-
Novel multiplex PCR reveals multiple trypanosomatid species infecting North American bumble bees (Hymenoptera: Apidae: Bombus).J Invertebr Pathol. 2018 Mar;153:147-155. doi: 10.1016/j.jip.2018.03.009. Epub 2018 Mar 14. J Invertebr Pathol. 2018. PMID: 29550403
-
The trypanosomatid (Kinetoplastida: Trypanosomatidae) parasites in bees: A review on their environmental circulation, impacts and implications.Curr Res Insect Sci. 2025 Jan 21;7:100106. doi: 10.1016/j.cris.2025.100106. eCollection 2025. Curr Res Insect Sci. 2025. PMID: 39925747 Free PMC article. Review.
-
Effects of invasive parasites on bumble bee declines.Conserv Biol. 2011 Aug;25(4):662-71. doi: 10.1111/j.1523-1739.2011.01707.x. Conserv Biol. 2011. PMID: 21771075 Review.
Cited by
-
Genomes of Endotrypanum monterogeii from Panama and Zelonia costaricensis from Brazil: Expansion of Multigene Families in Leishmaniinae Parasites That Are Close Relatives of Leishmania spp.Pathogens. 2023 Nov 30;12(12):1409. doi: 10.3390/pathogens12121409. Pathogens. 2023. PMID: 38133293 Free PMC article.
-
Molecular and Functional Characteristics of DNA Polymerase Beta-Like Enzymes From Trypanosomatids.Front Cell Infect Microbiol. 2021 Aug 5;11:670564. doi: 10.3389/fcimb.2021.670564. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34422676 Free PMC article. Review.
-
The evolution of trypanosomatid taxonomy.Parasit Vectors. 2017 Jun 8;10(1):287. doi: 10.1186/s13071-017-2204-7. Parasit Vectors. 2017. PMID: 28595622 Free PMC article. Review.
-
Pathogens Spillover from Honey Bees to Other Arthropods.Pathogens. 2021 Aug 17;10(8):1044. doi: 10.3390/pathogens10081044. Pathogens. 2021. PMID: 34451508 Free PMC article. Review.
-
The power and promise of applying genomics to honey bee health.Curr Opin Insect Sci. 2015 Aug 1;10:124-132. doi: 10.1016/j.cois.2015.03.007. Curr Opin Insect Sci. 2015. PMID: 26273565 Free PMC article.
References
-
- Langridge D, McGhee R (1967) Crithidia mellificae nsp an acidophilic trypanosomatide of honey bee Apis mellifera. J Protozool 14: 485–487. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

