Three groups of transposable elements with contrasting copy number dynamics and host responses in the maize (Zea mays ssp. mays) genome
- PMID: 24743518
- PMCID: PMC3990487
- DOI: 10.1371/journal.pgen.1004298
Three groups of transposable elements with contrasting copy number dynamics and host responses in the maize (Zea mays ssp. mays) genome
Abstract
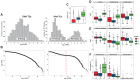
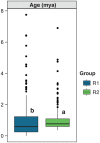
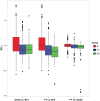
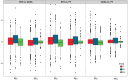
Most angiosperm nuclear DNA is repetitive and derived from silenced transposable elements (TEs). TE silencing requires substantial resources from the plant host, including the production of small interfering RNAs (siRNAs). Thus, the interaction between TEs and siRNAs is a critical aspect of both the function and the evolution of plant genomes. Yet the co-evolutionary dynamics between these two entities remain poorly characterized. Here we studied the organization of TEs within the maize (Zea mays ssp mays) genome, documenting that TEs fall within three groups based on the class and copy numbers. These groups included DNA elements, low copy RNA elements and higher copy RNA elements. The three groups varied statistically in characteristics that included length, location, age, siRNA expression and 24:22 nucleotide (nt) siRNA targeting ratios. In addition, the low copy retroelements encompassed a set of TEs that had previously been shown to decrease expression within a 24 nt siRNA biogenesis mutant (mop1). To investigate the evolutionary dynamics of the three groups, we estimated their abundance in two landraces, one with a genome similar in size to that of the maize reference and the other with a 30% larger genome. For all three accessions, we assessed TE abundance as well as 22 nt and 24 nt siRNA content within leaves. The high copy number retroelements are under targeted similarly by siRNAs among accessions, appear to be born of a rapid bust of activity, and may be currently transpositionally dead or limited. In contrast, the lower copy number group of retrolements are targeted more dynamically and have had a long and ongoing history of transposition in the maize genome.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Tenaillon MI, Hollister JD, Gaut BS (2010) A triptych of the evolution of plant transposable elements. Trends Plant Sci 15: 471–478. - PubMed
-
- Leonardo TE, Nuzhdin SV (2002) Intracellular battlegrounds: conflict and cooperation between transposable elements. Genet Res 80: 155–161. - PubMed
-
- Lisch D (2009) Epigenetic regulation of transposable elements in plants. Annu Rev Plant Biol 60: 43–66. - PubMed
-
- Axtell MJ (2013) Classification and Comparison of Small RNAs from Plants. Annu Rev Plant Biol 64: 137–159. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

