Analysis of molecular cytogenetic alteration in rhabdomyosarcoma by array comparative genomic hybridization
- PMID: 24743780
- PMCID: PMC3990535
- DOI: 10.1371/journal.pone.0094924
Analysis of molecular cytogenetic alteration in rhabdomyosarcoma by array comparative genomic hybridization
Abstract
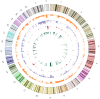
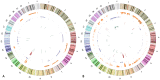
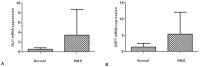
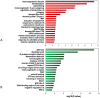
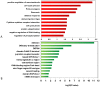
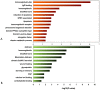
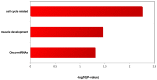
Rhabdomyosarcoma (RMS) is the most common pediatric soft tissue sarcoma with poor prognosis. The genetic etiology of RMS remains largely unclear underlying its development and progression. To reveal novel genes more precisely and new therapeutic targets associated with RMS, we used high-resolution array comparative genomic hybridization (aCGH) to explore tumor-associated copy number variations (CNVs) and genes in RMS. We confirmed several important genes by quantitative real-time polymerase chain reaction (QRT-PCR). We then performed bioinformatics-based functional enrichment analysis for genes located in the genomic regions with CNVs. In addition, we identified miRNAs located in the corresponding amplification and deletion regions and performed miRNA functional enrichment analysis. aCGH analyses revealed that all RMS showed specific gains and losses. The amplification regions were 12q13.12, 12q13.3, and 12q13.3-q14.1. The deletion regions were 1p21.1, 2q14.1, 5q13.2, 9p12, and 9q12. The recurrent regions with gains were 12q13.3, 12q13.3-q14.1, 12q14.1, and 17q25.1. The recurrent regions with losses were 9p12-p11.2, 10q11.21-q11.22, 14q32.33, 16p11.2, and 22q11.1. The mean mRNA level of GLI1 in RMS was 6.61-fold higher than that in controls (p = 0.0477) by QRT-PCR. Meanwhile, the mean mRNA level of GEFT in RMS samples was 3.92-fold higher than that in controls (p = 0.0354). Bioinformatic analysis showed that genes were enriched in functions such as immunoglobulin domain, induction of apoptosis, and defensin. Proto-oncogene functions were involved in alveolar RMS. miRNAs that located in the amplified regions in RMS tend to be enriched in oncogenic activity (miR-24 and miR-27a). In conclusion, this study identified a number of CNVs in RMS and functional analyses showed enrichment for genes and miRNAs located in these CNVs regions. These findings may potentially help the identification of novel biomarkers and/or drug targets implicated in diagnosis of and targeted therapy for RMS.
Conflict of interest statement
Figures

References
-
- Christopher D, Krisjnan U, Fredrik M (2002) World Health Organization classification of tumours. Pathology and genetics of tumours of soft tissue and bone.
-
- Barr FG (2001) Gene fusions involving PAX and FOX family members in alveolar rhabdomyosarcoma. Oncogene 20: 5736–5746. - PubMed
-
- Solinas-Toldo S, Lampel S, Stilgenbauer S, Nickolenko J, Benner A, et al. (1997) Matrix-based comparative genomic hybridization: biochips to screen for genomic imbalances. Genes, chromosomes and cancer 20: 399–407. - PubMed
-
- Pinkel D, Segraves R, Sudar D, Clark S, Poole I, et al. (1998) High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nature genetics 20: 207–211. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

