Evaluating the impact of genotype errors on rare variant tests of association
- PMID: 24744770
- PMCID: PMC3978329
- DOI: 10.3389/fgene.2014.00062
Evaluating the impact of genotype errors on rare variant tests of association
Abstract
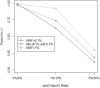
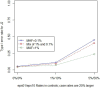
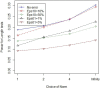
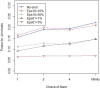
The new class of rare variant tests has usually been evaluated assuming perfect genotype information. In reality, rare variant genotypes may be incorrect, and so rare variant tests should be robust to imperfect data. Errors and uncertainty in SNP genotyping are already known to dramatically impact statistical power for single marker tests on common variants and, in some cases, inflate the type I error rate. Recent results show that uncertainty in genotype calls derived from sequencing reads are dependent on several factors, including read depth, calling algorithm, number of alleles present in the sample, and the frequency at which an allele segregates in the population. We have recently proposed a general framework for the evaluation and investigation of rare variant tests of association, classifying most rare variant tests into one of two broad categories (length or joint tests). We use this framework to relate factors affecting genotype uncertainty to the power and type I error rate of rare variant tests. We find that non-differential genotype errors (an error process that occurs independent of phenotype) decrease power, with larger decreases for extremely rare variants, and for the common homozygote to heterozygote error. Differential genotype errors (an error process that is associated with phenotype status), lead to inflated type I error rates which are more likely to occur at sites with more common homozygote to heterozygote errors than vice versa. Finally, our work suggests that certain rare variant tests and study designs may be more robust to the inclusion of genotype errors. Further work is needed to directly integrate genotype calling algorithm decisions, study costs and test statistic choices to provide comprehensive design and analysis advice which appropriately accounts for the impact of genotype errors.
Keywords: SKAT; dosage; gene-based; genotype uncertainty; misclassification.
Figures
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

