Changes in REDD1, REDD2, and atrogene mRNA expression are prevented in skeletal muscle fixed in a stretched position during hindlimb immobilization
- PMID: 24744910
- PMCID: PMC3966240
- DOI: 10.1002/phy2.246
Changes in REDD1, REDD2, and atrogene mRNA expression are prevented in skeletal muscle fixed in a stretched position during hindlimb immobilization
Abstract
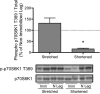
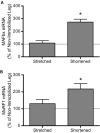
Immobilized skeletal muscle fixed in a shortened position displays disuse atrophy, whereas when fixed in a stretched position it does not (Goldspink, D. F. (1977) J Physiol 264, 267-282). Although significant advances have been made in our understanding of mechanisms involved in development of atrophy in muscle fixed in a shortened position, little is known about why mass is maintained when muscle is immobilized in a stretched position. In the present study, we hypothesized that skeletal muscle immobilized in a stretched position would be protected from gene expression changes known to be associated with disuse atrophy. To test the hypothesis, male Sprague-Dawley rats were anesthetized using isoflurane and subjected to unilateral hindlimb immobilization for 3 days with the soleus fixed in either a shortened or stretched position. All comparisons were made to the contralateral nonimmobilized muscle. Soleus immobilized in a shortened position exhibited disuse atrophy, attenuated rates of protein synthesis, attenuated mTORC1 signaling, and induced expression of genes for REDD1, REDD2, MAFbx, and MuRF1. In contrast, immobilization of the soleus in a stretched position prevented these changes as it exhibited no difference in muscle mass, rates of protein synthesis, mTORC1 signaling, or expression of genes encoding REDD1, MAFbx or MuRF1, with REDD2 expression being reduced compared to control. In conclusion, fixed muscle length plays a major role in immobilization-induced skeletal muscle atrophy whereby placing muscle in a shortened position leads to induction of gene expression for REDD1, REDD2, and atrogenes.
Keywords: Casting; Ddit4; Ddit4l; disuse atrophy; mTORC1.
Figures


Similar articles
-
Emerging role for regulated in development and DNA damage 1 (REDD1) in the regulation of skeletal muscle metabolism.Am J Physiol Endocrinol Metab. 2016 Jul 1;311(1):E157-74. doi: 10.1152/ajpendo.00059.2016. Epub 2016 May 17. Am J Physiol Endocrinol Metab. 2016. PMID: 27189933 Free PMC article. Review.
-
REDD2 expression in rat skeletal muscle correlates with nutrient-induced activation of mTORC1: responses to aging, immobilization, and remobilization.Am J Physiol Endocrinol Metab. 2015 Jan 15;308(2):E122-9. doi: 10.1152/ajpendo.00341.2014. Epub 2014 Nov 18. Am J Physiol Endocrinol Metab. 2015. PMID: 25406262 Free PMC article.
-
The mTORC1 signaling repressors REDD1/2 are rapidly induced and activation of p70S6K1 by leucine is defective in skeletal muscle of an immobilized rat hindlimb.Am J Physiol Endocrinol Metab. 2013 Jan 15;304(2):E229-36. doi: 10.1152/ajpendo.00409.2012. Epub 2012 Nov 27. Am J Physiol Endocrinol Metab. 2013. PMID: 23193052 Free PMC article.
-
Repressors of mTORC1 act to blunt the anabolic response to feeding in the soleus muscle of a cast-immobilized mouse hindlimb.Physiol Rep. 2018 Oct;6(20):e13891. doi: 10.14814/phy2.13891. Physiol Rep. 2018. PMID: 30338657 Free PMC article.
-
Signaling mechanisms involved in disuse muscle atrophy.Med Hypotheses. 2007;69(2):310-21. doi: 10.1016/j.mehy.2006.11.043. Epub 2007 Mar 21. Med Hypotheses. 2007. PMID: 17376604 Review.
Cited by
-
Endurance exercise induces REDD1 expression and transiently decreases mTORC1 signaling in rat skeletal muscle.Physiol Rep. 2014 Dec 24;2(12):e12254. doi: 10.14814/phy2.12254. Print 2014 Dec 1. Physiol Rep. 2014. PMID: 25539833 Free PMC article.
-
RNA sequencing reveals a slow to fast muscle fiber type transition after olanzapine infusion in rats.PLoS One. 2015 Apr 20;10(4):e0123966. doi: 10.1371/journal.pone.0123966. eCollection 2015. PLoS One. 2015. PMID: 25893406 Free PMC article.
-
Loss of REDD1 augments the rate of the overload-induced increase in muscle mass.Am J Physiol Regul Integr Comp Physiol. 2016 Sep 1;311(3):R545-57. doi: 10.1152/ajpregu.00159.2016. Epub 2016 Jul 27. Am J Physiol Regul Integr Comp Physiol. 2016. PMID: 27465734 Free PMC article.
-
RNA processing in skeletal muscle biology and disease.Transcription. 2019 Feb;10(1):1-20. doi: 10.1080/21541264.2018.1558677. Epub 2019 Jan 15. Transcription. 2019. PMID: 30556762 Free PMC article. Review.
-
Emerging role for regulated in development and DNA damage 1 (REDD1) in the regulation of skeletal muscle metabolism.Am J Physiol Endocrinol Metab. 2016 Jul 1;311(1):E157-74. doi: 10.1152/ajpendo.00059.2016. Epub 2016 May 17. Am J Physiol Endocrinol Metab. 2016. PMID: 27189933 Free PMC article. Review.
References
-
- Agata N., Sasai N., Inoue‐Miyazu M., Kawakami K., Hayakawa K., Kobayashi K. 2009. Repetitive stretch suppresses denervation‐induced atrophy of soleus muscle in rats. Muscle Nerve; 39:456-462 - PubMed
-
- Bajotto G., Sato Y., Kitaura Y., Shimomura Y. 2011. Effect of branched‐chain amino acid supplementation during unloading on regulatory components of protein synthesis in atrophied soleus muscles. Eur. J. Appl. Physiol.; 111:1815-1828 - PubMed
-
- Baptista I. L., Leal M. L., Artioli G. G., Aoki M. S., Fiamoncini J., Turri A. O. 2010. Leucine attenuates skeletal muscle wasting via inhibition of ubiquitin ligases. Muscle Nerve; 41:800-808 - PubMed
-
- Bodine S. C., Latres E., Baumhueter S., Lai V. K., Nunez L., Clarke B. A. 2001. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science; 294:1704-1708 - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

