C. elegans RNA-binding protein GLD-1 recognizes its multiple targets using sequence, context, and structural information to repress translation
- PMID: 24744981
- PMCID: PMC3988121
- DOI: 10.4161/worm.26548
C. elegans RNA-binding protein GLD-1 recognizes its multiple targets using sequence, context, and structural information to repress translation
Abstract
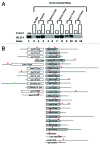
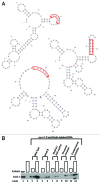
Caenorhabditis elegans GLD-1, a maxi-KH motif containing RNA-binding protein, has various functions mainly during female germ cell development, suggesting that it likely controls the expression of a selective group of maternal mRNAs. To gain an insight into how GLD-1 specifically recognizes these mRNA targets, we identified 38 biochemically proven GLD-1 binding regions from multiple mRNA targets that are among over 100 putative targets co-immunoprecipitated with GLD-1. The sequence information of these regions revealed three over-represented and phylogenetically conserved sequence motifs. We found that two of the motifs, one of which is novel, are important for GLD-1 binding in several GLD-1 binding regions but not in other regions. Further analyses indicate that the importance of one of the sequence motifs is dependent on two aspects: (1) surrounding sequence information, likely acting as an accessory feature for GLD-1 to efficiently select the sequence motif and (2) RNA secondary structural environment where the sequence motif resides, which likely provides "binding-site accessibility" for GLD-1 to effectively recognize its targets. Our data suggest some mRNAs recruit GLD-1 by a distinct mechanism, which involves more than one sequence motif that needs to be embedded in the correct context and structural environment.
Keywords: GLD-1; binding site accessibility; context dependency; mRNA targets; sequence motif.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources
