Maximizing the power of principal-component analysis of correlated phenotypes in genome-wide association studies
- PMID: 24746957
- PMCID: PMC4067564
- DOI: 10.1016/j.ajhg.2014.03.016
Maximizing the power of principal-component analysis of correlated phenotypes in genome-wide association studies
Abstract
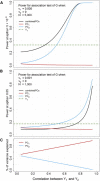
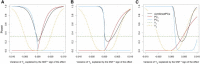
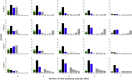
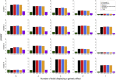
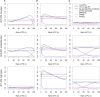
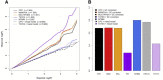
Many human traits are highly correlated. This correlation can be leveraged to improve the power of genetic association tests to identify markers associated with one or more of the traits. Principal component analysis (PCA) is a useful tool that has been widely used for the multivariate analysis of correlated variables. PCA is usually applied as a dimension reduction method: the few top principal components (PCs) explaining most of total trait variance are tested for association with a predictor of interest, and the remaining components are not analyzed. In this study we review the theoretical basis of PCA and describe the behavior of PCA when testing for association between a SNP and correlated traits. We then use simulation to compare the power of various PCA-based strategies when analyzing up to 100 correlated traits. We show that contrary to widespread practice, testing only the top PCs often has low power, whereas combining signal across all PCs can have greater power. This power gain is primarily due to increased power to detect genetic variants with opposite effects on positively correlated traits and variants that are exclusively associated with a single trait. Relative to other methods, the combined-PC approach has close to optimal power in all scenarios considered while offering more flexibility and more robustness to potential confounders. Finally, we apply the proposed PCA strategy to the genome-wide association study of five correlated coagulation traits where we identify two candidate SNPs that were not found by the standard approach.
Copyright © 2014 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

