Hematopoietic overexpression of FOG1 does not affect B-cells but reduces the number of circulating eosinophils
- PMID: 24747299
- PMCID: PMC3991581
- DOI: 10.1371/journal.pone.0092836
Hematopoietic overexpression of FOG1 does not affect B-cells but reduces the number of circulating eosinophils
Abstract
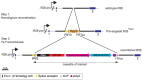
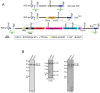
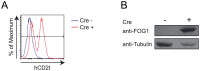
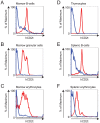
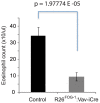
We have identified expression of the gene encoding the transcriptional coactivator FOG-1 (Friend of GATA-1; Zfpm1, Zinc finger protein multitype 1) in B lymphocytes. We found that FOG-1 expression is directly or indirectly dependent on the B cell-specific coactivator OBF-1 and that it is modulated during B cell development: expression is observed in early but not in late stages of B cell development. To directly test in vivo the role of FOG-1 in B lymphocytes, we developed a novel embryonic stem cell recombination system. For this, we combined homologous recombination with the FLP recombinase activity to rapidly generate embryonic stem cell lines carrying a Cre-inducible transgene at the Rosa26 locus. Using this system, we successfully generated transgenic mice where FOG-1 is conditionally overexpressed in mature B-cells or in the entire hematopoietic system. While overexpression of FOG-1 in B cells did not significantly affect B cell development or function, we found that enforced expression of FOG-1 throughout all hematopoietic lineages led to a reduction in the number of circulating eosinophils, confirming and extending to mammals the known function of FOG-1 in this lineage.
Conflict of interest statement
Figures



References
-
- Matthias P, Rolink AG (2005) Transcriptional networks in developing and mature B cells. Nature reviews Immunology 5: 497–508. - PubMed
-
- Cantor AB, Orkin SH (2005) Coregulation of GATA factors by the Friend of GATA (FOG) family of multitype zinc finger proteins. Seminars in cell & developmental biology 16: 117–128. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

