CasA mediates Cas3-catalyzed target degradation during CRISPR RNA-guided interference
- PMID: 24748111
- PMCID: PMC4020112
- DOI: 10.1073/pnas.1405079111
CasA mediates Cas3-catalyzed target degradation during CRISPR RNA-guided interference
Abstract
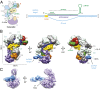
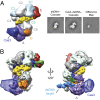
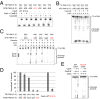
In bacteria, the clustered regularly interspaced short palindromic repeats (CRISPR)-associated (Cas) DNA-targeting complex Cascade (CRISPR-associated complex for antiviral defense) uses CRISPR RNA (crRNA) guides to bind complementary DNA targets at sites adjacent to a trinucleotide signature sequence called the protospacer adjacent motif (PAM). The Cascade complex then recruits Cas3, a nuclease-helicase that catalyzes unwinding and cleavage of foreign double-stranded DNA (dsDNA) bearing a sequence matching that of the crRNA. Cascade comprises the CasA-E proteins and one crRNA, forming a structure that binds and unwinds dsDNA to form an R loop in which the target strand of the DNA base pairs with the 32-nt RNA guide sequence. Single-particle electron microscopy reconstructions of dsDNA-bound Cascade with and without Cas3 reveal that Cascade positions the PAM-proximal end of the DNA duplex at the CasA subunit and near the site of Cas3 association. The finding that the DNA target and Cas3 colocalize with CasA implicates this subunit in a key target-validation step during DNA interference. We show biochemically that base pairing of the PAM region is unnecessary for target binding but critical for Cas3-mediated degradation. In addition, the L1 loop of CasA, previously implicated in PAM recognition, is essential for Cas3 activation following target binding by Cascade. Together, these data show that the CasA subunit of Cascade functions as an essential partner of Cas3 by recognizing DNA target sites and positioning Cas3 adjacent to the PAM to ensure cleavage.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Structural basis for promiscuous PAM recognition in type I-E Cascade from E. coli.Nature. 2016 Feb 25;530(7591):499-503. doi: 10.1038/nature16995. Epub 2016 Feb 10. Nature. 2016. PMID: 26863189 Free PMC article.
-
Molecular insights into DNA interference by CRISPR-associated nuclease-helicase Cas3.Proc Natl Acad Sci U S A. 2014 Nov 18;111(46):16359-64. doi: 10.1073/pnas.1410806111. Epub 2014 Nov 3. Proc Natl Acad Sci U S A. 2014. PMID: 25368186 Free PMC article.
-
Cas3-Derived Target DNA Degradation Fragments Fuel Primed CRISPR Adaptation.Mol Cell. 2016 Sep 1;63(5):852-64. doi: 10.1016/j.molcel.2016.07.011. Epub 2016 Aug 18. Mol Cell. 2016. PMID: 27546790
-
Fitting CRISPR-associated Cas3 into the helicase family tree.Curr Opin Struct Biol. 2014 Feb;24:106-14. doi: 10.1016/j.sbi.2014.01.001. Epub 2014 Jan 28. Curr Opin Struct Biol. 2014. PMID: 24480304 Free PMC article. Review.
-
Unity among the diverse RNA-guided CRISPR-Cas interference mechanisms.J Biol Chem. 2024 Jun;300(6):107295. doi: 10.1016/j.jbc.2024.107295. Epub 2024 Apr 18. J Biol Chem. 2024. PMID: 38641067 Free PMC article. Review.
Cited by
-
Structural insights into the inactivation of CRISPR-Cas systems by diverse anti-CRISPR proteins.BMC Biol. 2018 Mar 19;16(1):32. doi: 10.1186/s12915-018-0504-9. BMC Biol. 2018. PMID: 29554913 Free PMC article. Review.
-
Systematic prediction of genes functionally linked to CRISPR-Cas systems by gene neighborhood analysis.Proc Natl Acad Sci U S A. 2018 Jun 5;115(23):E5307-E5316. doi: 10.1073/pnas.1803440115. Epub 2018 May 21. Proc Natl Acad Sci U S A. 2018. PMID: 29784811 Free PMC article.
-
Type I CRISPR-Cas targets endogenous genes and regulates virulence to evade mammalian host immunity.Cell Res. 2016 Dec;26(12):1273-1287. doi: 10.1038/cr.2016.135. Epub 2016 Nov 18. Cell Res. 2016. PMID: 27857054 Free PMC article.
-
CRISPR-Based Genome Editing Tools: An Accelerator in Crop Breeding for a Changing Future.Int J Mol Sci. 2023 May 11;24(10):8623. doi: 10.3390/ijms24108623. Int J Mol Sci. 2023. PMID: 37239967 Free PMC article. Review.
-
Structural basis for assembly of non-canonical small subunits into type I-C Cascade.Nat Commun. 2020 Nov 23;11(1):5931. doi: 10.1038/s41467-020-19785-8. Nat Commun. 2020. PMID: 33230133 Free PMC article.
References
-
- Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482(7385):331–338. - PubMed
-
- Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315(5819):1709–1712. - PubMed
-
- Garneau JE, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468(7320):67–71. - PubMed
-
- Mojica FJ, Díez-Villaseñor C, García-Martínez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155(Pt 3):733–740. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

