Uncovering the protein lysine and arginine methylation network in Arabidopsis chloroplasts
- PMID: 24748391
- PMCID: PMC3991674
- DOI: 10.1371/journal.pone.0095512
Uncovering the protein lysine and arginine methylation network in Arabidopsis chloroplasts
Abstract
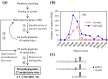
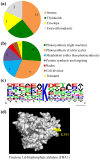
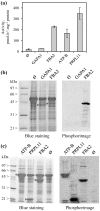
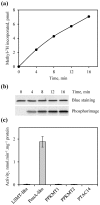
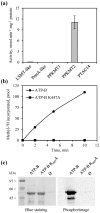
Post-translational modification of proteins by the addition of methyl groups to the side chains of Lys and Arg residues is proposed to play important roles in many cellular processes. In plants, identification of non-histone methylproteins at a cellular or subcellular scale is still missing. To gain insights into the extent of this modification in chloroplasts we used a bioinformatics approach to identify protein methyltransferases targeted to plastids and set up a workflow to specifically identify Lys and Arg methylated proteins from proteomic data used to produce the Arabidopsis chloroplast proteome. With this approach we could identify 31 high-confidence Lys and Arg methylation sites from 23 chloroplastic proteins, of which only two were previously known to be methylated. These methylproteins are split between the stroma, thylakoids and envelope sub-compartments. They belong to essential metabolic processes, including photosynthesis, and to the chloroplast biogenesis and maintenance machinery (translation, protein import, division). Also, the in silico identification of nine protein methyltransferases that are known or predicted to be targeted to plastids provided a foundation to build the enzymes/substrates relationships that govern methylation in chloroplasts. Thereby, using in vitro methylation assays with chloroplast stroma as a source of methyltransferases we confirmed the methylation sites of two targets, plastid ribosomal protein L11 and the β-subunit of ATP synthase. Furthermore, a biochemical screening of recombinant chloroplastic protein Lys methyltransferases allowed us to identify the enzymes involved in the modification of these substrates. The present study provides a useful resource to build the methyltransferases/methylproteins network and to elucidate the role of protein methylation in chloroplast biology.
Conflict of interest statement
Figures

Similar articles
-
Dual Targeting of the Protein Methyltransferase PrmA Contributes to Both Chloroplastic and Mitochondrial Ribosomal Protein L11 Methylation in Arabidopsis.Plant Cell Physiol. 2015 Sep;56(9):1697-710. doi: 10.1093/pcp/pcv098. Epub 2015 Jun 26. Plant Cell Physiol. 2015. PMID: 26116422
-
Proteins of diverse function and subcellular location are lysine acetylated in Arabidopsis.Plant Physiol. 2011 Apr;155(4):1779-90. doi: 10.1104/pp.110.171595. Epub 2011 Feb 10. Plant Physiol. 2011. PMID: 21311031 Free PMC article.
-
N4-methylcytidine ribosomal RNA methylation in chloroplasts is crucial for chloroplast function, development, and abscisic acid response in Arabidopsis.J Integr Plant Biol. 2021 Mar;63(3):570-582. doi: 10.1111/jipb.13009. Epub 2020 Sep 30. J Integr Plant Biol. 2021. PMID: 32876986
-
Chloroplast proteomics and the compartmentation of plastidial isoprenoid biosynthetic pathways.Mol Plant. 2009 Nov;2(6):1154-80. doi: 10.1093/mp/ssp088. Epub 2009 Nov 12. Mol Plant. 2009. PMID: 19969518 Review.
-
Chloroplast Proteases: Updates on Proteolysis within and across Suborganellar Compartments.Plant Physiol. 2016 Aug;171(4):2280-93. doi: 10.1104/pp.16.00330. Epub 2016 Jun 10. Plant Physiol. 2016. PMID: 27288365 Free PMC article. Review.
Cited by
-
Knocking Out Chloroplastic Aldolases/Rubisco Lysine Methyltransferase Enhances Biomass Accumulation in Nannochloropsis oceanica under High-Light Stress.Int J Mol Sci. 2024 Mar 28;25(7):3756. doi: 10.3390/ijms25073756. Int J Mol Sci. 2024. PMID: 38612566 Free PMC article.
-
Large Scale Mass Spectrometry-based Identifications of Enzyme-mediated Protein Methylation Are Subject to High False Discovery Rates.Mol Cell Proteomics. 2016 Mar;15(3):989-1006. doi: 10.1074/mcp.M115.055384. Epub 2015 Dec 23. Mol Cell Proteomics. 2016. PMID: 26699799 Free PMC article.
-
Protein Methylation and Translation: Role of Lysine Modification on the Function of Yeast Elongation Factor 1A.Biochemistry. 2019 Dec 10;58(49):4997-5010. doi: 10.1021/acs.biochem.9b00818. Epub 2019 Dec 2. Biochemistry. 2019. PMID: 31738538 Free PMC article.
-
Engineering Triterpene and Methylated Triterpene Production in Plants Provides Biochemical and Physiological Insights into Terpene Metabolism.Plant Physiol. 2016 Feb;170(2):702-16. doi: 10.1104/pp.15.01548. Epub 2015 Nov 24. Plant Physiol. 2016. PMID: 26603654 Free PMC article.
-
Root morphogenic pathways in Eucalyptus grandis are modified by the activity of protein arginine methyltransferases.BMC Plant Biol. 2017 Mar 9;17(1):62. doi: 10.1186/s12870-017-1010-x. BMC Plant Biol. 2017. PMID: 28279165 Free PMC article.
References
-
- Egorova KS, Olenkina OM, Olenina LV (2010) Lysine methylation of nonhistone proteins is a way to regulate their stability and function. Biochemistry (Mosc) 75: 535–548. - PubMed
-
- Huang J, Berger SL (2008) The emerging field of dynamic lysine methylation of non-histone proteins. Curr Opin Genet Dev 18: 152–158. - PubMed
-
- Zhang L, Ma H (2012) Complex evolutionary history and diverse domain organization of SET proteins suggest divergent regulatory interactions. New Phytol 195: 248–263. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

