Streptococcus mutans extracellular DNA is upregulated during growth in biofilms, actively released via membrane vesicles, and influenced by components of the protein secretion machinery
- PMID: 24748612
- PMCID: PMC4054167
- DOI: 10.1128/JB.01493-14
Streptococcus mutans extracellular DNA is upregulated during growth in biofilms, actively released via membrane vesicles, and influenced by components of the protein secretion machinery
Abstract
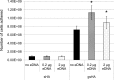
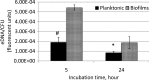
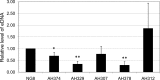
Streptococcus mutans, a major etiological agent of human dental caries, lives primarily on the tooth surface in biofilms. Limited information is available concerning the extracellular DNA (eDNA) as a scaffolding matrix in S. mutans biofilms. This study demonstrates that S. mutans produces eDNA by multiple avenues, including lysis-independent membrane vesicles. Unlike eDNAs from cell lysis that were abundant and mainly concentrated around broken cells or cell debris with floating open ends, eDNAs produced via the lysis-independent pathway appeared scattered but in a structured network under scanning electron microscopy. Compared to eDNA production of planktonic cultures, eDNA production in 5- and 24-h biofilms was increased by >3- and >1.6-fold, respectively. The addition of DNase I to growth medium significantly reduced biofilm formation. In an in vitro adherence assay, added chromosomal DNA alone had a limited effect on S. mutans adherence to saliva-coated hydroxylapatite beads, but in conjunction with glucans synthesized using purified glucosyltransferase B, the adherence was significantly enhanced. Deletion of sortase A, the transpeptidase that covalently couples multiple surface-associated proteins to the cell wall peptidoglycan, significantly reduced eDNA in both planktonic and biofilm cultures. Sortase A deficiency did not have a significant effect on membrane vesicle production; however, the protein profile of the mutant membrane vesicles was significantly altered, including reduction of adhesin P1 and glucan-binding proteins B and C. Relative to the wild type, deficiency of protein secretion and membrane protein insertion machinery components, including Ffh, YidC1, and YidC2, also caused significant reductions in eDNA.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures





Similar articles
-
Glycosyltransferase-Mediated Biofilm Matrix Dynamics and Virulence of Streptococcus mutans.Appl Environ Microbiol. 2019 Feb 20;85(5):e02247-18. doi: 10.1128/AEM.02247-18. Print 2019 Mar 1. Appl Environ Microbiol. 2019. PMID: 30578260 Free PMC article.
-
Competence-Stimulating-Peptide-Dependent Localized Cell Death and Extracellular DNA Production in Streptococcus mutans Biofilms.Appl Environ Microbiol. 2020 Nov 10;86(23):e02080-20. doi: 10.1128/AEM.02080-20. Print 2020 Nov 10. Appl Environ Microbiol. 2020. PMID: 32948520 Free PMC article.
-
Effects of Complex DNA and MVs with GTF Extracted from Streptococcus mutans on the Oral Biofilm.Molecules. 2019 Aug 28;24(17):3131. doi: 10.3390/molecules24173131. Molecules. 2019. PMID: 31466323 Free PMC article.
-
Streptococcus mutans-derived extracellular matrix in cariogenic oral biofilms.Front Cell Infect Microbiol. 2015 Feb 13;5:10. doi: 10.3389/fcimb.2015.00010. eCollection 2015. Front Cell Infect Microbiol. 2015. PMID: 25763359 Free PMC article. Review.
-
The role of extracellular DNA in the formation, architecture, stability, and treatment of bacterial biofilms.Biotechnol Bioeng. 2021 Jun;118(6):2129-2141. doi: 10.1002/bit.27760. Epub 2021 Mar 27. Biotechnol Bioeng. 2021. PMID: 33748946 Free PMC article. Review.
Cited by
-
Unravelling the DNA sequences carried by Streptomyces coelicolor membrane vesicles.Sci Rep. 2022 Oct 5;12(1):16651. doi: 10.1038/s41598-022-21002-z. Sci Rep. 2022. PMID: 36198712 Free PMC article.
-
Effects of Novel Dental Composites on Streptococcus mutans Biofilms.J Funct Biomater. 2023 Dec 29;15(1):13. doi: 10.3390/jfb15010013. J Funct Biomater. 2023. PMID: 38248680 Free PMC article.
-
Gram-Positive Bacterial Extracellular Vesicles and Their Impact on Health and Disease.Front Microbiol. 2018 Jul 9;9:1502. doi: 10.3389/fmicb.2018.01502. eCollection 2018. Front Microbiol. 2018. PMID: 30038605 Free PMC article.
-
Bacterial extracellular vesicles as bioactive nanocarriers for drug delivery: Advances and perspectives.Bioact Mater. 2021 Dec 17;14:169-181. doi: 10.1016/j.bioactmat.2021.12.006. eCollection 2022 Aug. Bioact Mater. 2021. PMID: 35310361 Free PMC article. Review.
-
Phage-Mediated Explosive Cell Lysis Induces the Formation of a Different Type of O-IMV in Shewanella vesiculosa M7T.Front Microbiol. 2021 Oct 8;12:713669. doi: 10.3389/fmicb.2021.713669. eCollection 2021. Front Microbiol. 2021. PMID: 34690958 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

